Please be patient as the page loads
|
JARD2_HUMAN
|
||||||
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
JARD2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
JARD2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.965229 (rank : 2) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | 2.26708e-35 (rank : 3) | NC score | 0.666710 (rank : 7) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
JAD1C_HUMAN
|
||||||
| θ value | 6.59618e-35 (rank : 4) | NC score | 0.673388 (rank : 5) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
JAD1C_MOUSE
|
||||||
| θ value | 6.59618e-35 (rank : 5) | NC score | 0.674897 (rank : 4) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
JAD1D_MOUSE
|
||||||
| θ value | 1.46948e-34 (rank : 6) | NC score | 0.676284 (rank : 3) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JAD1D_HUMAN
|
||||||
| θ value | 5.584e-34 (rank : 7) | NC score | 0.672705 (rank : 6) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JHD3D_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 8) | NC score | 0.637364 (rank : 8) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6B0I6, Q9NT41, Q9NW76 | Gene names | JMJD2D, JHDM3D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
JHD3C_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 9) | NC score | 0.549677 (rank : 10) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
JHD3D_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 10) | NC score | 0.637122 (rank : 9) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3U2K5, Q2M1G7, Q8BI19 | Gene names | Jmjd2d, Jhdm3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
JHD3B_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 11) | NC score | 0.543935 (rank : 12) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3C_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 12) | NC score | 0.544765 (rank : 11) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H3R0, O94877, Q2M3M0, Q5JUC9, Q5VYJ2, Q5VYJ3 | Gene names | JMJD2C, GASC1, JHDM3C, KIAA0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C) (Gene amplified in squamous cell carcinoma 1 protein) (GASC-1 protein). | |||||
|
JHD3B_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 13) | NC score | 0.543255 (rank : 13) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3A_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 14) | NC score | 0.541097 (rank : 14) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3A_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 15) | NC score | 0.540888 (rank : 15) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 16) | NC score | 0.288326 (rank : 18) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 17) | NC score | 0.252153 (rank : 19) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
ARI3A_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 18) | NC score | 0.352480 (rank : 16) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62431 | Gene names | Arid3a, Dri1, Dril1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (Dead ringer-like protein 1) (B-cell regulator of IgH transcription) (Bright). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 19) | NC score | 0.205899 (rank : 21) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
ARI3A_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 20) | NC score | 0.340026 (rank : 17) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99856, Q8IZA7 | Gene names | ARID3A, DRIL1, DRX, E2FBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (B-cell regulator of IgH transcription) (Bright) (E2F-binding protein 1). | |||||
|
CD2L5_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 21) | NC score | 0.021866 (rank : 135) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
CD2L5_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 22) | NC score | 0.022152 (rank : 134) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14004, Q6DKQ9, Q75MH4, Q75MH5, Q96JN4, Q9H4A0, Q9H4A1, Q9UDR4 | Gene names | CDC2L5, CDC2L, CHED, KIAA1791 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5) (Cholinesterase-related cell division controller). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 23) | NC score | 0.110134 (rank : 36) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 24) | NC score | 0.224351 (rank : 20) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
TM16H_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.047865 (rank : 93) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.061191 (rank : 71) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
PHAR2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 27) | NC score | 0.052286 (rank : 84) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
PRP4B_MOUSE
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.018730 (rank : 141) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
RBMX2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.039365 (rank : 98) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R0F5, Q3TI42 | Gene names | Rbmx2 | |||
|
Domain Architecture |
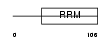
|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
ENL_HUMAN
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.059303 (rank : 74) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.057740 (rank : 77) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 32) | NC score | 0.043416 (rank : 96) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 33) | NC score | 0.069073 (rank : 63) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ZN192_HUMAN
|
||||||
| θ value | 0.21417 (rank : 34) | NC score | 0.001607 (rank : 173) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15776, Q4VAR1, Q4VAR2, Q4VAR3, Q9H4T1 | Gene names | ZNF192 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 192 (LD5-1). | |||||
|
ZN694_HUMAN
|
||||||
| θ value | 0.21417 (rank : 35) | NC score | 0.004899 (rank : 168) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q63HK3, Q6ZN77 | Gene names | ZNF694 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 694. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 36) | NC score | 0.031190 (rank : 113) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 37) | NC score | 0.180614 (rank : 23) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
ARI5B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 38) | NC score | 0.192361 (rank : 22) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.114891 (rank : 35) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
CCNT2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.035166 (rank : 106) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
RBMX2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.046242 (rank : 95) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y388, Q5JY82, Q9Y3I8 | Gene names | RBMX2 | |||
|
Domain Architecture |
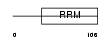
|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
GP158_HUMAN
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.039166 (rank : 99) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
PCX1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.037154 (rank : 103) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.038329 (rank : 100) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.028857 (rank : 117) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
NFRKB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.033335 (rank : 108) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.048377 (rank : 92) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.060701 (rank : 72) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
DVL1L_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.023502 (rank : 131) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54792 | Gene names | DVL1L1, DVL, DVL1 | |||
|
Domain Architecture |
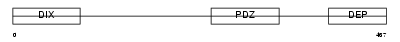
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1-like (Dishevelled- 1-like) (DSH homolog 1-like). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.029649 (rank : 116) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.064822 (rank : 65) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.033673 (rank : 107) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
FBX40_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.023696 (rank : 130) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UH90, Q9ULM5 | Gene names | FBXO40, FBX40, KIAA1195 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 40 (Muscle disease-related protein). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.016091 (rank : 146) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
BCLF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.030257 (rank : 114) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
CAR10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.014160 (rank : 152) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BWT7, Q14CQ8, Q5TFG6, Q9UGR5, Q9UGR6, Q9Y3H0 | Gene names | CARD10, CARMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (CARD-containing MAGUK protein 3) (Carma 3). | |||||
|
DNMBP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.012417 (rank : 153) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6TXD4, Q6ZQ05, Q8CEW8, Q8R0Y2 | Gene names | Dnmbp, Kiaa1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
DVL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.021819 (rank : 136) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14640 | Gene names | DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
HRG_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.032828 (rank : 109) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04196 | Gene names | HRG | |||
|
Domain Architecture |
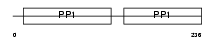
|
|||||
| Description | Histidine-rich glycoprotein precursor (Histidine-proline-rich glycoprotein) (HPRG). | |||||
|
MARE2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.022188 (rank : 133) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R001, Q3UF61, Q8BLZ6, Q8BYR6, Q8C177, Q8K109 | Gene names | Mapre2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein RP/EB family member 2 (APC-binding protein EB2) (End-binding protein 2) (EB2). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.051815 (rank : 87) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.027796 (rank : 119) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TNR1B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.015860 (rank : 147) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20333, Q16042, Q6YI29, Q9UIH1 | Gene names | TNFRSF1B, TNFBR, TNFR2 | |||
|
Domain Architecture |
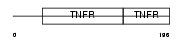
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor) (CD120b antigen) (Etanercept) [Contains: Tumor necrosis factor receptor superfamily member 1b, membrane form; Tumor necrosis factor-binding protein 2 (TBPII) (TBP- 2)]. | |||||
|
UBP53_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.031830 (rank : 111) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q70EK8, Q68DA5, Q8WVQ5, Q9P2J7 | Gene names | USP53, KIAA1350 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53). | |||||
|
ZN350_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.000532 (rank : 175) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9GZX5, Q9HAQ4 | Gene names | ZNF350, ZBRK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 350 (Zinc-finger protein ZBRK1) (Zinc finger and BRCA1-interacting protein with a KRAB domain 1) (KRAB zinc finger protein ZFQR). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.019448 (rank : 137) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.012214 (rank : 155) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
PJA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.019241 (rank : 139) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55176, Q8CFU2, Q99MJ1, Q99MJ2, Q99MJ3, Q9DB04 | Gene names | Pja1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.051961 (rank : 86) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.014833 (rank : 150) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
MECP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.024233 (rank : 127) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2D6 | Gene names | Mecp2 | |||
|
Domain Architecture |
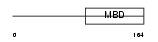
|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.036067 (rank : 105) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.028245 (rank : 118) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.026539 (rank : 121) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
SUHW3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.004919 (rank : 167) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P3Y5, Q6ZPM2, Q8K145 | Gene names | Suhw3, Kiaa1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
TAU_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.025035 (rank : 125) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.014975 (rank : 149) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.031616 (rank : 112) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
ADDA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.025699 (rank : 123) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35611, Q13734, Q14729, Q16156, Q9UJB6 | Gene names | ADD1, ADDA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.011692 (rank : 156) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.009519 (rank : 159) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
IKBE_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.008078 (rank : 163) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.046815 (rank : 94) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.003113 (rank : 171) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
TR150_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.036502 (rank : 104) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.025950 (rank : 122) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
AF9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.039486 (rank : 97) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
BBX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.010400 (rank : 157) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
ELOA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.019153 (rank : 140) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CB77, Q80VB2, Q9R0Q5 | Gene names | Tceb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.009040 (rank : 161) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.029669 (rank : 115) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.026954 (rank : 120) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.024161 (rank : 128) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
NUFP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.018639 (rank : 142) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5F2E7, Q3TCE2, Q3V195, Q80TF1 | Gene names | Nufip2, Kiaa1321 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 2 (FMRP- interacting protein 2) (82 kDa FMRP-interacting protein) (82-FIP). | |||||
|
TMC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.025136 (rank : 124) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R4P4 | Gene names | Tmc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
DIP2A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.009510 (rank : 160) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWT5, Q8CHI0 | Gene names | Dip2a, Dip2, Kiaa0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.006456 (rank : 166) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
HB2Q_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.001332 (rank : 174) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13763 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DP(W2) beta chain precursor (SB-2-beta). | |||||
|
MARE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.016819 (rank : 145) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15555, Q9UQ33 | Gene names | MAPRE2, RP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein RP/EB family member 2 (APC-binding protein EB2) (End-binding protein 2) (EB2). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.057481 (rank : 78) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.016896 (rank : 144) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.024033 (rank : 129) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.012282 (rank : 154) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TOP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.019426 (rank : 138) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11387, O43256, Q12855, Q12856, Q15610, Q5TFY3, Q9UJN0 | Gene names | TOP1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
BAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.008996 (rank : 162) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92560, Q6LEM0, Q7Z5E8 | Gene names | BAP1, KIAA0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Cerebral protein 6). | |||||
|
BCOR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.007866 (rank : 164) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
DAAM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.007388 (rank : 165) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80U19, Q810J5 | Gene names | Daam2, Kiaa0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
ETV3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.004536 (rank : 169) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
HORN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.032824 (rank : 110) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
MMTA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.014347 (rank : 151) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BU76, Q6P5Y0, Q6ZTZ6, Q6ZWA6, Q8IZH3 | Gene names | MMTAG2, C1orf35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 (hMMTAG2). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.038160 (rank : 101) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
OMGP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.002143 (rank : 172) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q63912 | Gene names | Omg, Omgp | |||
|
Domain Architecture |
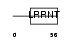
|
|||||
| Description | Oligodendrocyte-myelin glycoprotein precursor. | |||||
|
PRP40_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.024276 (rank : 126) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.015177 (rank : 148) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.037804 (rank : 102) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
TBR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.003939 (rank : 170) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64336 | Gene names | Tbr1 | |||
|
Domain Architecture |
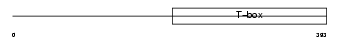
|
|||||
| Description | T-brain-1 protein (T-box brain protein 1) (TBR-1) (TES-56). | |||||
|
TOP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.017407 (rank : 143) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q04750 | Gene names | Top1, Top-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
UBP53_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.023177 (rank : 132) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
US6NL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.009593 (rank : 158) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
AF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.132796 (rank : 28) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.131833 (rank : 29) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.148060 (rank : 26) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.100077 (rank : 40) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.099198 (rank : 41) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.088796 (rank : 46) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.098310 (rank : 43) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.081321 (rank : 57) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.088167 (rank : 47) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.054403 (rank : 81) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.067263 (rank : 64) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
BRD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.123589 (rank : 30) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.134493 (rank : 27) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.104032 (rank : 38) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
CHD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.054996 (rank : 80) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.051405 (rank : 88) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.052147 (rank : 85) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.050634 (rank : 91) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.051221 (rank : 90) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
DPF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.083073 (rank : 54) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
DPF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.083597 (rank : 52) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
JADE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.116049 (rank : 34) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.116102 (rank : 33) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.104200 (rank : 37) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.101261 (rank : 39) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JADE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.117353 (rank : 32) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.120003 (rank : 31) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.088117 (rank : 48) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.087373 (rank : 49) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.093105 (rank : 44) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.059976 (rank : 73) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.056777 (rank : 79) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.064208 (rank : 68) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.063658 (rank : 69) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.054374 (rank : 82) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.083489 (rank : 53) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.085603 (rank : 51) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.090533 (rank : 45) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.086990 (rank : 50) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.078420 (rank : 59) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.079303 (rank : 58) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.064298 (rank : 67) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.064640 (rank : 66) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.161262 (rank : 25) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.162275 (rank : 24) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.058293 (rank : 76) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
REQU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.082465 (rank : 55) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.082448 (rank : 56) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.098806 (rank : 42) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.058321 (rank : 75) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.051249 (rank : 89) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.053780 (rank : 83) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
UHRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.076650 (rank : 60) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
UHRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.071817 (rank : 61) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
UHRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.069750 (rank : 62) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
UHRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.063502 (rank : 70) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
JARD2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
JARD2_MOUSE
|
||||||
| NC score | 0.965229 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
JAD1D_MOUSE
|
||||||
| NC score | 0.676284 (rank : 3) | θ value | 1.46948e-34 (rank : 6) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JAD1C_MOUSE
|
||||||
| NC score | 0.674897 (rank : 4) | θ value | 6.59618e-35 (rank : 5) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
JAD1C_HUMAN
|
||||||
| NC score | 0.673388 (rank : 5) | θ value | 6.59618e-35 (rank : 4) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
JAD1D_HUMAN
|
||||||
| NC score | 0.672705 (rank : 6) | θ value | 5.584e-34 (rank : 7) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.666710 (rank : 7) | θ value | 2.26708e-35 (rank : 3) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
JHD3D_HUMAN
|
||||||
| NC score | 0.637364 (rank : 8) | θ value | 4.02038e-16 (rank : 8) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6B0I6, Q9NT41, Q9NW76 | Gene names | JMJD2D, JHDM3D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
JHD3D_MOUSE
|
||||||
| NC score | 0.637122 (rank : 9) | θ value | 1.52774e-15 (rank : 10) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3U2K5, Q2M1G7, Q8BI19 | Gene names | Jmjd2d, Jhdm3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
JHD3C_MOUSE
|
||||||
| NC score | 0.549677 (rank : 10) | θ value | 1.52774e-15 (rank : 9) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
JHD3C_HUMAN
|
||||||
| NC score | 0.544765 (rank : 11) | θ value | 2.60593e-15 (rank : 12) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H3R0, O94877, Q2M3M0, Q5JUC9, Q5VYJ2, Q5VYJ3 | Gene names | JMJD2C, GASC1, JHDM3C, KIAA0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C) (Gene amplified in squamous cell carcinoma 1 protein) (GASC-1 protein). | |||||
|
JHD3B_MOUSE
|
||||||
| NC score | 0.543935 (rank : 12) | θ value | 2.60593e-15 (rank : 11) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3B_HUMAN
|
||||||
| NC score | 0.543255 (rank : 13) | θ value | 1.29331e-14 (rank : 13) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3A_MOUSE
|
||||||
| NC score | 0.541097 (rank : 14) | θ value | 2.43908e-13 (rank : 14) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3A_HUMAN
|
||||||
| NC score | 0.540888 (rank : 15) | θ value | 3.18553e-13 (rank : 15) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
ARI3A_MOUSE
|
||||||
| NC score | 0.352480 (rank : 16) | θ value | 7.1131e-05 (rank : 18) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62431 | Gene names | Arid3a, Dri1, Dril1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (Dead ringer-like protein 1) (B-cell regulator of IgH transcription) (Bright). | |||||
|
ARI3A_HUMAN
|
||||||
| NC score | 0.340026 (rank : 17) | θ value | 0.000786445 (rank : 20) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99856, Q8IZA7 | Gene names | ARID3A, DRIL1, DRX, E2FBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (B-cell regulator of IgH transcription) (Bright) (E2F-binding protein 1). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.288326 (rank : 18) | θ value | 8.40245e-06 (rank : 16) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.252153 (rank : 19) | θ value | 1.43324e-05 (rank : 17) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.224351 (rank : 20) | θ value | 0.0330416 (rank : 24) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.205899 (rank : 21) | θ value | 0.000786445 (rank : 19) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
ARI5B_MOUSE
|
||||||
| NC score | 0.192361 (rank : 22) | θ value | 0.279714 (rank : 38) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.180614 (rank : 23) | θ value | 0.279714 (rank : 37) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.162275 (rank : 24) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.161262 (rank : 25) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.148060 (rank : 26) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.134493 (rank : 27) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
AF10_HUMAN
|
||||||
| NC score | 0.132796 (rank : 28) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_MOUSE
|
||||||
| NC score | 0.131833 (rank : 29) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
BRD1_HUMAN
|
||||||
| NC score | 0.123589 (rank : 30) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
JADE3_MOUSE
|
||||||
| NC score | 0.120003 (rank : 31) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE3_HUMAN
|
||||||
| NC score | 0.117353 (rank : 32) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE1_MOUSE
|
||||||
| NC score | 0.116102 (rank : 33) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_HUMAN
|
||||||
| NC score | 0.116049 (rank : 34) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.114891 (rank : 35) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.110134 (rank : 36) | θ value | 0.0148317 (rank : 23) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
JADE2_HUMAN
|
||||||
| NC score | 0.104200 (rank : 37) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.104032 (rank : 38) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.101261 (rank : 39) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.100077 (rank : 40) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.099198 (rank : 41) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.098806 (rank : 42) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.098310 (rank : 43) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.093105 (rank : 44) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.090533 (rank : 45) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.088796 (rank : 46) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.088167 (rank : 47) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.088117 (rank : 48) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.087373 (rank : 49) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.086990 (rank : 50) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.085603 (rank : 51) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
DPF1_MOUSE
|
||||||
| NC score | 0.083597 (rank : 52) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.083489 (rank : 53) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
DPF1_HUMAN
|
||||||
| NC score | 0.083073 (rank : 54) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
REQU_HUMAN
|
||||||
| NC score | 0.082465 (rank : 55) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| NC score | 0.082448 (rank : 56) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.081321 (rank : 57) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.079303 (rank : 58) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.078420 (rank : 59) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
UHRF1_HUMAN
|
||||||
| NC score | 0.076650 (rank : 60) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
UHRF1_MOUSE
|
||||||
| NC score | 0.071817 (rank : 61) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
UHRF2_HUMAN
|
||||||
| NC score | 0.069750 (rank : 62) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.069073 (rank : 63) | θ value | 0.163984 (rank : 33) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.067263 (rank : 64) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.064822 (rank : 65) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.064640 (rank : 66) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.064298 (rank : 67) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.064208 (rank : 68) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.063658 (rank : 69) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
UHRF2_MOUSE
|
||||||
| NC score | 0.063502 (rank : 70) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.061191 (rank : 71) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.060701 (rank : 72) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.059976 (rank : 73) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.059303 (rank : 74) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.058321 (rank : 75) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.058293 (rank : 76) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.057740 (rank : 77) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.057481 (rank : 78) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.056777 (rank : 79) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.054996 (rank : 80) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.054403 (rank : 81) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.054374 (rank : 82) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.053780 (rank : 83) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
PHAR2_HUMAN
|
||||||
| NC score | 0.052286 (rank : 84) | θ value | 0.0961366 (rank : 27) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.052147 (rank : 85) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.051961 (rank : 86) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.051815 (rank : 87) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.051405 (rank : 88) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.051249 (rank : 89) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.051221 (rank : 90) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.050634 (rank : 91) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.048377 (rank : 92) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TM16H_HUMAN
|
||||||
| NC score | 0.047865 (rank : 93) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.046815 (rank : 94) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
RBMX2_HUMAN
|
||||||
| NC score | 0.046242 (rank : 95) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y388, Q5JY82, Q9Y3I8 | Gene names | RBMX2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.043416 (rank : 96) | θ value | 0.163984 (rank : 32) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.039486 (rank : 97) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
RBMX2_MOUSE
|
||||||
| NC score | 0.039365 (rank : 98) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R0F5, Q3TI42 | Gene names | Rbmx2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.039166 (rank : 99) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.038329 (rank : 100) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.038160 (rank : 101) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.037804 (rank : 102) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
PCX1_HUMAN
|
||||||
| NC score | 0.037154 (rank : 103) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.036502 (rank : 104) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.036067 (rank : 105) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
CCNT2_HUMAN
|
||||||
| NC score | 0.035166 (rank : 106) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.033673 (rank : 107) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
NFRKB_HUMAN
|
||||||
| NC score | 0.033335 (rank : 108) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
HRG_HUMAN
|
||||||
| NC score | 0.032828 (rank : 109) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04196 | Gene names | HRG | |||
|
Domain Architecture |

|
|||||
| Description | Histidine-rich glycoprotein precursor (Histidine-proline-rich glycoprotein) (HPRG). | |||||
|
HORN_MOUSE
|
||||||
| NC score | 0.032824 (rank : 110) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
UBP53_HUMAN
|
||||||
| NC score | 0.031830 (rank : 111) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q70EK8, Q68DA5, Q8WVQ5, Q9P2J7 | Gene names | USP53, KIAA1350 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.031616 (rank : 112) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.031190 (rank : 113) | θ value | 0.279714 (rank : 36) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
BCLF1_MOUSE
|
||||||
| NC score | 0.030257 (rank : 114) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.029669 (rank : 115) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.029649 (rank : 116) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.028857 (rank : 117) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.028245 (rank : 118) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.027796 (rank : 119) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.026954 (rank : 120) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.026539 (rank : 121) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.025950 (rank : 122) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
ADDA_HUMAN
|
||||||
| NC score | 0.025699 (rank : 123) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35611, Q13734, Q14729, Q16156, Q9UJB6 | Gene names | ADD1, ADDA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
TMC2_MOUSE
|
||||||
| NC score | 0.025136 (rank : 124) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R4P4 | Gene names | Tmc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.025035 (rank : 125) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.024276 (rank : 126) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
MECP2_MOUSE
|
||||||
| NC score | 0.024233 (rank : 127) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2D6 | Gene names | Mecp2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.024161 (rank : 128) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.024033 (rank : 129) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
FBX40_HUMAN
|
||||||
| NC score | 0.023696 (rank : 130) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UH90, Q9ULM5 | Gene names | FBXO40, FBX40, KIAA1195 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 40 (Muscle disease-related protein). | |||||
|
DVL1L_HUMAN
|
||||||
| NC score | 0.023502 (rank : 131) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54792 | Gene names | DVL1L1, DVL, DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1-like (Dishevelled- 1-like) (DSH homolog 1-like). | |||||
|
UBP53_MOUSE
|
||||||
| NC score | 0.023177 (rank : 132) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
MARE2_MOUSE
|
||||||
| NC score | 0.022188 (rank : 133) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R001, Q3UF61, Q8BLZ6, Q8BYR6, Q8C177, Q8K109 | Gene names | Mapre2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein RP/EB family member 2 (APC-binding protein EB2) (End-binding protein 2) (EB2). | |||||
|
CD2L5_HUMAN
|
||||||
| NC score | 0.022152 (rank : 134) | θ value | 0.00175202 (rank : 22) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14004, Q6DKQ9, Q75MH4, Q75MH5, Q96JN4, Q9H4A0, Q9H4A1, Q9UDR4 | Gene names | CDC2L5, CDC2L, CHED, KIAA1791 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5) (Cholinesterase-related cell division controller). | |||||
|
CD2L5_MOUSE
|
||||||
| NC score | 0.021866 (rank : 135) | θ value | 0.00134147 (rank : 21) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
DVL1_HUMAN
|
||||||
| NC score | 0.021819 (rank : 136) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14640 | Gene names | DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.019448 (rank : 137) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
TOP1_HUMAN
|
||||||
| NC score | 0.019426 (rank : 138) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11387, O43256, Q12855, Q12856, Q15610, Q5TFY3, Q9UJN0 | Gene names | TOP1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
PJA1_MOUSE
|
||||||
| NC score | 0.019241 (rank : 139) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55176, Q8CFU2, Q99MJ1, Q99MJ2, Q99MJ3, Q9DB04 | Gene names | Pja1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-). | |||||
|
ELOA1_MOUSE
|
||||||
| NC score | 0.019153 (rank : 140) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CB77, Q80VB2, Q9R0Q5 | Gene names | Tceb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
PRP4B_MOUSE
|
||||||
| NC score | 0.018730 (rank : 141) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
NUFP2_MOUSE
|
||||||
| NC score | 0.018639 (rank : 142) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5F2E7, Q3TCE2, Q3V195, Q80TF1 | Gene names | Nufip2, Kiaa1321 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 2 (FMRP- interacting protein 2) (82 kDa FMRP-interacting protein) (82-FIP). | |||||
|
TOP1_MOUSE
|
||||||
| NC score | 0.017407 (rank : 143) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q04750 | Gene names | Top1, Top-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.016896 (rank : 144) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
MARE2_HUMAN
|
||||||
| NC score | 0.016819 (rank : 145) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15555, Q9UQ33 | Gene names | MAPRE2, RP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein RP/EB family member 2 (APC-binding protein EB2) (End-binding protein 2) (EB2). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.016091 (rank : 146) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
TNR1B_HUMAN
|
||||||
| NC score | 0.015860 (rank : 147) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20333, Q16042, Q6YI29, Q9UIH1 | Gene names | TNFRSF1B, TNFBR, TNFR2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor) (CD120b antigen) (Etanercept) [Contains: Tumor necrosis factor receptor superfamily member 1b, membrane form; Tumor necrosis factor-binding protein 2 (TBPII) (TBP- 2)]. | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.015177 (rank : 148) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.014975 (rank : 149) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.014833 (rank : 150) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
MMTA2_HUMAN
|
||||||
| NC score | 0.014347 (rank : 151) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BU76, Q6P5Y0, Q6ZTZ6, Q6ZWA6, Q8IZH3 | Gene names | MMTAG2, C1orf35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 (hMMTAG2). | |||||
|
CAR10_HUMAN
|
||||||
| NC score | 0.014160 (rank : 152) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BWT7, Q14CQ8, Q5TFG6, Q9UGR5, Q9UGR6, Q9Y3H0 | Gene names | CARD10, CARMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (CARD-containing MAGUK protein 3) (Carma 3). | |||||
|
DNMBP_MOUSE
|
||||||
| NC score | 0.012417 (rank : 153) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6TXD4, Q6ZQ05, Q8CEW8, Q8R0Y2 | Gene names | Dnmbp, Kiaa1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.012282 (rank : 154) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.012214 (rank : 155) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.011692 (rank : 156) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BBX_MOUSE
|
||||||
| NC score | 0.010400 (rank : 157) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
US6NL_MOUSE
|
||||||
| NC score | 0.009593 (rank : 158) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.009519 (rank : 159) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
DIP2A_MOUSE
|
||||||
| NC score | 0.009510 (rank : 160) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWT5, Q8CHI0 | Gene names | Dip2a, Dip2, Kiaa0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.009040 (rank : 161) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
BAP1_HUMAN
|
||||||
| NC score | 0.008996 (rank : 162) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92560, Q6LEM0, Q7Z5E8 | Gene names | BAP1, KIAA0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Cerebral protein 6). | |||||
|
IKBE_MOUSE
|
||||||
| NC score | 0.008078 (rank : 163) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
BCOR_MOUSE
|
||||||
| NC score | 0.007866 (rank : 164) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
DAAM2_MOUSE
|
||||||
| NC score | 0.007388 (rank : 165) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80U19, Q810J5 | Gene names | Daam2, Kiaa0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.006456 (rank : 166) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
SUHW3_MOUSE
|
||||||
| NC score | 0.004919 (rank : 167) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P3Y5, Q6ZPM2, Q8K145 | Gene names | Suhw3, Kiaa1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
ZN694_HUMAN
|
||||||
| NC score | 0.004899 (rank : 168) | θ value | 0.21417 (rank : 35) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q63HK3, Q6ZN77 | Gene names | ZNF694 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 694. | |||||
|
ETV3_MOUSE
|
||||||
| NC score | 0.004536 (rank : 169) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
TBR1_MOUSE
|
||||||
| NC score | 0.003939 (rank : 170) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64336 | Gene names | Tbr1 | |||
|
Domain Architecture |

|
|||||
| Description | T-brain-1 protein (T-box brain protein 1) (TBR-1) (TES-56). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.003113 (rank : 171) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
OMGP_MOUSE
|
||||||
| NC score | 0.002143 (rank : 172) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q63912 | Gene names | Omg, Omgp | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte-myelin glycoprotein precursor. | |||||
|
ZN192_HUMAN
|
||||||
| NC score | 0.001607 (rank : 173) | θ value | 0.21417 (rank : 34) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15776, Q4VAR1, Q4VAR2, Q4VAR3, Q9H4T1 | Gene names | ZNF192 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 192 (LD5-1). | |||||
|
HB2Q_HUMAN
|
||||||
| NC score | 0.001332 (rank : 174) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13763 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DP(W2) beta chain precursor (SB-2-beta). | |||||
|
ZN350_HUMAN
|
||||||
| NC score | 0.000532 (rank : 175) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9GZX5, Q9HAQ4 | Gene names | ZNF350, ZBRK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 350 (Zinc-finger protein ZBRK1) (Zinc finger and BRCA1-interacting protein with a KRAB domain 1) (KRAB zinc finger protein ZFQR). | |||||