Please be patient as the page loads
|
JHD3B_HUMAN
|
||||||
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
JHD3B_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3B_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989189 (rank : 2) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3C_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.971122 (rank : 6) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9H3R0, O94877, Q2M3M0, Q5JUC9, Q5VYJ2, Q5VYJ3 | Gene names | JMJD2C, GASC1, JHDM3C, KIAA0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C) (Gene amplified in squamous cell carcinoma 1 protein) (GASC-1 protein). | |||||
|
JHD3C_MOUSE
|
||||||
| θ value | 4.68096e-182 (rank : 4) | NC score | 0.973812 (rank : 3) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
JHD3A_HUMAN
|
||||||
| θ value | 6.12769e-174 (rank : 5) | NC score | 0.972314 (rank : 4) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3A_MOUSE
|
||||||
| θ value | 8.00302e-174 (rank : 6) | NC score | 0.971855 (rank : 5) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3D_MOUSE
|
||||||
| θ value | 6.35588e-163 (rank : 7) | NC score | 0.879211 (rank : 7) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3U2K5, Q2M1G7, Q8BI19 | Gene names | Jmjd2d, Jhdm3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
JHD3D_HUMAN
|
||||||
| θ value | 1.19867e-161 (rank : 8) | NC score | 0.877437 (rank : 8) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6B0I6, Q9NT41, Q9NW76 | Gene names | JMJD2D, JHDM3D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | 4.72714e-33 (rank : 9) | NC score | 0.616019 (rank : 13) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
JAD1D_HUMAN
|
||||||
| θ value | 5.77852e-31 (rank : 10) | NC score | 0.619079 (rank : 12) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JAD1C_HUMAN
|
||||||
| θ value | 7.54701e-31 (rank : 11) | NC score | 0.606841 (rank : 20) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
JAD1C_MOUSE
|
||||||
| θ value | 7.54701e-31 (rank : 12) | NC score | 0.607785 (rank : 18) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
JAD1D_MOUSE
|
||||||
| θ value | 2.86786e-30 (rank : 13) | NC score | 0.606852 (rank : 19) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | 7.06379e-29 (rank : 14) | NC score | 0.606767 (rank : 21) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
BRD1_HUMAN
|
||||||
| θ value | 1.02001e-27 (rank : 15) | NC score | 0.592954 (rank : 23) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | 3.62785e-25 (rank : 16) | NC score | 0.572381 (rank : 27) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 1.16704e-23 (rank : 17) | NC score | 0.573497 (rank : 26) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JADE2_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 18) | NC score | 0.604412 (rank : 22) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
AF17_HUMAN
|
||||||
| θ value | 2.59989e-23 (rank : 19) | NC score | 0.674451 (rank : 9) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
AF10_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 20) | NC score | 0.638522 (rank : 11) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_MOUSE
|
||||||
| θ value | 1.6852e-22 (rank : 21) | NC score | 0.640037 (rank : 10) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
JADE1_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 22) | NC score | 0.609521 (rank : 16) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_MOUSE
|
||||||
| θ value | 1.6852e-22 (rank : 23) | NC score | 0.608820 (rank : 17) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE3_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 24) | NC score | 0.612830 (rank : 14) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE3_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 25) | NC score | 0.612120 (rank : 15) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 26) | NC score | 0.580101 (rank : 25) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_MOUSE
|
||||||
| θ value | 1.38178e-16 (rank : 27) | NC score | 0.591673 (rank : 24) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
JARD2_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 28) | NC score | 0.546685 (rank : 28) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
JARD2_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 29) | NC score | 0.543255 (rank : 29) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 30) | NC score | 0.185925 (rank : 32) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 31) | NC score | 0.172586 (rank : 34) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 32) | NC score | 0.184720 (rank : 33) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 33) | NC score | 0.019812 (rank : 138) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PHF11_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 34) | NC score | 0.214080 (rank : 30) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 35) | NC score | 0.023051 (rank : 133) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 36) | NC score | 0.033997 (rank : 120) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 37) | NC score | 0.166472 (rank : 36) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
LBR_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 38) | NC score | 0.100361 (rank : 68) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14739, Q14740, Q53GU7, Q59FE6 | Gene names | LBR | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B receptor (Integral nuclear envelope inner membrane protein) (LMN2R). | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 39) | NC score | 0.145665 (rank : 44) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
DCC_MOUSE
|
||||||
| θ value | 0.163984 (rank : 40) | NC score | 0.007031 (rank : 170) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
LBR_MOUSE
|
||||||
| θ value | 0.163984 (rank : 41) | NC score | 0.096357 (rank : 74) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3U9G9, Q3TSW2, Q811V8, Q811V9, Q8BST3, Q8K2Y8, Q8VDM0, Q91YS5, Q91Z27 | Gene names | Lbr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lamin-B receptor (Integral nuclear envelope inner membrane protein). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 0.163984 (rank : 42) | NC score | 0.106470 (rank : 63) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 0.163984 (rank : 43) | NC score | 0.106749 (rank : 62) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 44) | NC score | 0.167406 (rank : 35) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.025227 (rank : 129) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
PHF7_MOUSE
|
||||||
| θ value | 0.47712 (rank : 46) | NC score | 0.191441 (rank : 31) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.022355 (rank : 135) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.005908 (rank : 175) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.015661 (rank : 143) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
BSCL2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.031474 (rank : 122) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
PHF13_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.056194 (rank : 111) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86YI8, Q8N551, Q9UJP2 | Gene names | PHF13 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 13. | |||||
|
PHF13_MOUSE
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.056253 (rank : 110) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K2W6 | Gene names | Phf13 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 13. | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.134159 (rank : 47) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RAVR1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.013739 (rank : 146) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CW46, Q5DTT6, Q811K0, Q8C3Z1, Q8CA14 | Gene names | Raver1, Kiaa1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
TNAP2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.020589 (rank : 136) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03169, Q86VI0 | Gene names | TNFAIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 2 (Primary response gene B94 protein). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.095083 (rank : 78) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
FBX46_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.018940 (rank : 140) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
PCGF6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.017490 (rank : 142) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYE7, Q5SYD1, Q5SYD6, Q96ID9, Q96SJ1 | Gene names | PCGF6, MBLR, RNF134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb group RING finger protein 6 (RING finger protein 134) (Mel18 and Bmi1-like RING finger). | |||||
|
PHF7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.149796 (rank : 42) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.029304 (rank : 124) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.010096 (rank : 158) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
APLP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.013650 (rank : 149) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.048803 (rank : 116) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
BSCL2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.029743 (rank : 123) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2E9, Q810B0, Q9JJC2, Q9JMF1 | Gene names | Bscl2, Gng3lg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein homolog). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.041513 (rank : 117) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
RANB3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.020020 (rank : 137) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CT10, Q3TIS4, Q3U2G4, Q3UYG7 | Gene names | Ranbp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
CCD45_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.011989 (rank : 154) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
K1333_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.104405 (rank : 65) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.013219 (rank : 151) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.026761 (rank : 128) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CI040_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.024364 (rank : 131) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCE4, Q8BJJ5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf40 homolog. | |||||
|
E41L3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.012901 (rank : 152) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.024604 (rank : 130) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.034886 (rank : 119) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.028185 (rank : 127) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
WDR24_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.023483 (rank : 132) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96S15, Q96GC7, Q9H0B7 | Gene names | WDR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 24. | |||||
|
WDR24_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.018378 (rank : 141) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFJ9 | Gene names | Wdr24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 24. | |||||
|
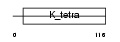
HKR3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | -0.003211 (rank : 184) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10074 | Gene names | HKR3 | |||
|
Domain Architecture |
|
|||||
| Description | Krueppel-related zinc finger protein 3 (Protein HKR3). | |||||
|
INSM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.002274 (rank : 179) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 683 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q01101 | Gene names | INSM1, IA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.038155 (rank : 118) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
OTU7A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.010393 (rank : 157) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
PHF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.157514 (rank : 40) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
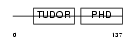
PHF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.158665 (rank : 39) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |
|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
PHF6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.105180 (rank : 64) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
PHF6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.097807 (rank : 71) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.007483 (rank : 167) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
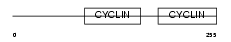
TF3B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.014082 (rank : 145) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92994, Q13223, Q5PR24, Q6IQ02, Q9HCW6, Q9HCW7, Q9HCW8 | Gene names | BRF1, BRF | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor IIIB 90 kDa subunit (TFIIIB90) (hTFIIIB90) (B- related factor 1) (BRF-1) (hBRF) (TATA box-binding protein-associated factor, RNA polymerase III, subunit 2) (TAF3B2). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.013652 (rank : 148) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
FARP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.006207 (rank : 174) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
KLF13_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | -0.002553 (rank : 182) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJZ6, Q9ESX3, Q9JHF8 | Gene names | Klf13, Bteb3, Fklf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 13 (Transcription factor BTEB3) (Basic transcription element-binding protein 3) (BTE-binding protein 3) (RANTES factor of late activated T-lymphocytes 1) (RFLAT-1) (Erythroid transcription factor FKLF-2). | |||||
|
MUS81_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.012399 (rank : 153) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91ZJ0, Q8R356 | Gene names | Mus81 | |||
|
Domain Architecture |

|
|||||
| Description | Crossover junction endonuclease MUS81 (EC 3.1.22.-). | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.053919 (rank : 114) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
PKN3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | -0.003069 (rank : 183) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
RAI2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.013628 (rank : 150) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y5P3, Q8N6X7 | Gene names | RAI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2. | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.004808 (rank : 177) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
FARP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.006767 (rank : 171) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
K1333_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.078553 (rank : 90) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.002753 (rank : 178) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
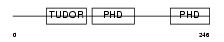
MTF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.142514 (rank : 46) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y483, Q9UES9, Q9UP40 | Gene names | MTF2 | |||
|
Domain Architecture |
|
|||||
| Description | Metal-response element-binding transcription factor 2 (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MTF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.146216 (rank : 43) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q02395, Q569Z8, Q6PG89, Q8BGP9, Q8BSJ7 | Gene names | Mtf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-response element-binding transcription factor 2 (Zinc-regulated factor 1) (ZiRF1) (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.001504 (rank : 180) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.028484 (rank : 126) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.096050 (rank : 76) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TRM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.013684 (rank : 147) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXH9, O76103, Q548Y5, Q8WVA6 | Gene names | TRMT1 | |||
|
Domain Architecture |

|
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.009206 (rank : 161) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.009638 (rank : 159) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.088989 (rank : 84) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.023023 (rank : 134) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.084851 (rank : 86) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
LCE2A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.019422 (rank : 139) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
NCOA7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.009513 (rank : 160) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6DFV7, Q3TAR1, Q3UNB3, Q66K05, Q6ZQM4, Q8BJ15, Q8BJ39 | Gene names | Ncoa7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7. | |||||
|
PO6F1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.007765 (rank : 164) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.098726 (rank : 69) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.029050 (rank : 125) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DGCR8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.007038 (rank : 169) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WYQ5, Q6DCB2, Q6MZE9, Q6Y2L0, Q96G39, Q96GP8, Q9H6L8, Q9H6T7, Q9NRW2 | Gene names | DGCR8, C22orf12, DGCRK6 | |||
|
Domain Architecture |

|
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8). | |||||
|
GNAS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.008010 (rank : 162) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5JWF2, O75684, O75685, Q5JW67, Q5JWF1, Q9NY42 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
IL2RB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.006739 (rank : 172) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14784 | Gene names | IL2RB | |||
|
Domain Architecture |
|
|||||
| Description | Interleukin-2 receptor beta chain precursor (IL-2 receptor) (P70-75) (p75) (High affinity IL-2 receptor subunit beta) (CD122 antigen). | |||||
|
JIP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.007629 (rank : 165) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
PAIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.007481 (rank : 168) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
PCGF6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.010900 (rank : 156) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99NA9 | Gene names | Pcgf6, Mblr, Rnf134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb group RING finger protein 6 (RING finger protein 134) (Mel18 and Bmi1-like RING finger). | |||||
|
PTN22_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.007519 (rank : 166) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
RNZ2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.006734 (rank : 173) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQ52, Q9HAS8, Q9HAS9, Q9NVT1 | Gene names | ELAC2, HPC2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc phosphodiesterase ELAC protein 2 (EC 3.1.26.11) (Ribonuclease Z 2) (RNase Z 2) (tRNase Z 2) (tRNA 3 endonuclease 2) (ElaC homolog protein 2) (Heredity prostate cancer protein 2). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.014569 (rank : 144) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.033685 (rank : 121) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TOPB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.007841 (rank : 163) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
TRM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.011877 (rank : 155) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TX08, Q3TBY4 | Gene names | Trmt1, D8Ertd812e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
WDR46_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.005824 (rank : 176) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15213, Q5STK5 | Gene names | WDR46, BING4, C6orf11 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
ZN447_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | -0.001850 (rank : 181) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.151293 (rank : 41) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.143017 (rank : 45) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.061914 (rank : 104) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.067043 (rank : 98) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
ARI3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.107473 (rank : 59) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99856, Q8IZA7 | Gene names | ARID3A, DRIL1, DRX, E2FBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (B-cell regulator of IgH transcription) (Bright) (E2F-binding protein 1). | |||||
|
ARI3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.110142 (rank : 55) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62431 | Gene names | Arid3a, Dri1, Dril1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (Dead ringer-like protein 1) (B-cell regulator of IgH transcription) (Bright). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.066407 (rank : 99) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
ARI5B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.068208 (rank : 96) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.122848 (rank : 50) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.120913 (rank : 51) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.131191 (rank : 48) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.109174 (rank : 57) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.107185 (rank : 60) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.101624 (rank : 67) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.097754 (rank : 72) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
BRD7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.056993 (rank : 109) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
BRD7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.055566 (rank : 112) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88665, Q3UQ56, Q3UU06, Q9CT78 | Gene names | Brd7, Bp75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein). | |||||
|
BRD9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.057964 (rank : 107) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H8M2, Q2XUS1, Q6UWU9, Q8IUS4, Q9H5Q5, Q9H7R9 | Gene names | BRD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9 (Rhabdomyosarcoma antigen MU-RMS- 40.8). | |||||
|
BRD9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.055420 (rank : 113) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UQU0, Q5FWH1, Q811F7 | Gene names | Brd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9. | |||||
|
CHD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.078247 (rank : 91) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.078061 (rank : 92) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.077650 (rank : 93) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
DPF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.107033 (rank : 61) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
DPF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.115873 (rank : 52) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
EPC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.052013 (rank : 115) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H2F5, Q5VW54, Q5VW56, Q5VW58, Q8NAQ4, Q8NE21, Q96LF4, Q96RR6, Q9H7T7 | Gene names | EPC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 1. | |||||
|
MYST3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.098323 (rank : 70) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.092534 (rank : 81) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.094358 (rank : 79) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.095841 (rank : 77) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.096927 (rank : 73) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.104228 (rank : 66) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.110652 (rank : 53) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.109518 (rank : 56) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.084809 (rank : 87) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.082935 (rank : 88) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.159021 (rank : 38) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.159024 (rank : 37) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.094175 (rank : 80) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.092345 (rank : 82) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.057473 (rank : 108) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.085444 (rank : 85) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
REQU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.110344 (rank : 54) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.108566 (rank : 58) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.125535 (rank : 49) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
TIF1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.065517 (rank : 101) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.063561 (rank : 102) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.062082 (rank : 103) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
TIF1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.066090 (rank : 100) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.058226 (rank : 106) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.059850 (rank : 105) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.067627 (rank : 97) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRI66_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.069294 (rank : 95) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
UHRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.081525 (rank : 89) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
UHRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.076673 (rank : 94) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
UHRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.089238 (rank : 83) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
UHRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.096269 (rank : 75) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
JHD3B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3B_MOUSE
|
||||||
| NC score | 0.989189 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3C_MOUSE
|
||||||
| NC score | 0.973812 (rank : 3) | θ value | 4.68096e-182 (rank : 4) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
JHD3A_HUMAN
|
||||||
| NC score | 0.972314 (rank : 4) | θ value | 6.12769e-174 (rank : 5) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3A_MOUSE
|
||||||
| NC score | 0.971855 (rank : 5) | θ value | 8.00302e-174 (rank : 6) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3C_HUMAN
|
||||||
| NC score | 0.971122 (rank : 6) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9H3R0, O94877, Q2M3M0, Q5JUC9, Q5VYJ2, Q5VYJ3 | Gene names | JMJD2C, GASC1, JHDM3C, KIAA0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C) (Gene amplified in squamous cell carcinoma 1 protein) (GASC-1 protein). | |||||
|
JHD3D_MOUSE
|
||||||
| NC score | 0.879211 (rank : 7) | θ value | 6.35588e-163 (rank : 7) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3U2K5, Q2M1G7, Q8BI19 | Gene names | Jmjd2d, Jhdm3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
JHD3D_HUMAN
|
||||||
| NC score | 0.877437 (rank : 8) | θ value | 1.19867e-161 (rank : 8) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6B0I6, Q9NT41, Q9NW76 | Gene names | JMJD2D, JHDM3D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.674451 (rank : 9) | θ value | 2.59989e-23 (rank : 19) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
AF10_MOUSE
|
||||||
| NC score | 0.640037 (rank : 10) | θ value | 1.6852e-22 (rank : 21) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_HUMAN
|
||||||
| NC score | 0.638522 (rank : 11) | θ value | 1.6852e-22 (rank : 20) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
JAD1D_HUMAN
|
||||||
| NC score | 0.619079 (rank : 12) | θ value | 5.77852e-31 (rank : 10) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.616019 (rank : 13) | θ value | 4.72714e-33 (rank : 9) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
JADE3_HUMAN
|
||||||
| NC score | 0.612830 (rank : 14) | θ value | 2.87452e-22 (rank : 24) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE3_MOUSE
|
||||||
| NC score | 0.612120 (rank : 15) | θ value | 3.75424e-22 (rank : 25) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE1_HUMAN
|
||||||
| NC score | 0.609521 (rank : 16) | θ value | 1.6852e-22 (rank : 22) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_MOUSE
|
||||||
| NC score | 0.608820 (rank : 17) | θ value | 1.6852e-22 (rank : 23) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JAD1C_MOUSE
|
||||||
| NC score | 0.607785 (rank : 18) | θ value | 7.54701e-31 (rank : 12) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
JAD1D_MOUSE
|
||||||
| NC score | 0.606852 (rank : 19) | θ value | 2.86786e-30 (rank : 13) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JAD1C_HUMAN
|
||||||
| NC score | 0.606841 (rank : 20) | θ value | 7.54701e-31 (rank : 11) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.606767 (rank : 21) | θ value | 7.06379e-29 (rank : 14) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
JADE2_HUMAN
|
||||||
| NC score | 0.604412 (rank : 22) | θ value | 1.99067e-23 (rank : 18) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
BRD1_HUMAN
|
||||||
| NC score | 0.592954 (rank : 23) | θ value | 1.02001e-27 (rank : 15) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.591673 (rank : 24) | θ value | 1.38178e-16 (rank : 27) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.580101 (rank : 25) | θ value | 1.38178e-16 (rank : 26) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.573497 (rank : 26) | θ value | 1.16704e-23 (rank : 17) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.572381 (rank : 27) | θ value | 3.62785e-25 (rank : 16) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
JARD2_MOUSE
|
||||||
| NC score | 0.546685 (rank : 28) | θ value | 5.25075e-16 (rank : 28) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
JARD2_HUMAN
|
||||||
| NC score | 0.543255 (rank : 29) | θ value | 1.29331e-14 (rank : 29) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
PHF11_HUMAN
|
||||||
| NC score | 0.214080 (rank : 30) | θ value | 0.0148317 (rank : 34) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
PHF7_MOUSE
|
||||||
| NC score | 0.191441 (rank : 31) | θ value | 0.47712 (rank : 46) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.185925 (rank : 32) | θ value | 0.00175202 (rank : 30) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.184720 (rank : 33) | θ value | 0.0113563 (rank : 32) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.172586 (rank : 34) | θ value | 0.00390308 (rank : 31) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.167406 (rank : 35) | θ value | 0.21417 (rank : 44) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.166472 (rank : 36) | θ value | 0.0563607 (rank : 37) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.159024 (rank : 37) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.159021 (rank : 38) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF1_MOUSE
|
||||||
| NC score | 0.158665 (rank : 39) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
PHF1_HUMAN
|
||||||
| NC score | 0.157514 (rank : 40) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.151293 (rank : 41) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
PHF7_HUMAN
|
||||||
| NC score | 0.149796 (rank : 42) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
MTF2_MOUSE
|
||||||
| NC score | 0.146216 (rank : 43) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q02395, Q569Z8, Q6PG89, Q8BGP9, Q8BSJ7 | Gene names | Mtf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-response element-binding transcription factor 2 (Zinc-regulated factor 1) (ZiRF1) (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.145665 (rank : 44) | θ value | 0.0961366 (rank : 39) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.143017 (rank : 45) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
MTF2_HUMAN
|
||||||
| NC score | 0.142514 (rank : 46) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y483, Q9UES9, Q9UP40 | Gene names | MTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Metal-response element-binding transcription factor 2 (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.134159 (rank : 47) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.131191 (rank : 48) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.125535 (rank : 49) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.122848 (rank : 50) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.120913 (rank : 51) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
DPF1_MOUSE
|
||||||
| NC score | 0.115873 (rank : 52) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.110652 (rank : 53) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
REQU_HUMAN
|
||||||
| NC score | 0.110344 (rank : 54) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
ARI3A_MOUSE
|
||||||
| NC score | 0.110142 (rank : 55) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62431 | Gene names | Arid3a, Dri1, Dril1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (Dead ringer-like protein 1) (B-cell regulator of IgH transcription) (Bright). | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.109518 (rank : 56) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.109174 (rank : 57) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
REQU_MOUSE
|
||||||
| NC score | 0.108566 (rank : 58) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
ARI3A_HUMAN
|
||||||
| NC score | 0.107473 (rank : 59) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99856, Q8IZA7 | Gene names | ARID3A, DRIL1, DRX, E2FBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (B-cell regulator of IgH transcription) (Bright) (E2F-binding protein 1). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.107185 (rank : 60) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
DPF1_HUMAN
|
||||||
| NC score | 0.107033 (rank : 61) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.106749 (rank : 62) | θ value | 0.163984 (rank : 43) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.106470 (rank : 63) | θ value | 0.163984 (rank : 42) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
PHF6_HUMAN
|
||||||
| NC score | 0.105180 (rank : 64) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
K1333_MOUSE
|
||||||
| NC score | 0.104405 (rank : 65) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.104228 (rank : 66) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.101624 (rank : 67) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
LBR_HUMAN
|
||||||
| NC score | 0.100361 (rank : 68) | θ value | 0.0563607 (rank : 38) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14739, Q14740, Q53GU7, Q59FE6 | Gene names | LBR | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B receptor (Integral nuclear envelope inner membrane protein) (LMN2R). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.098726 (rank : 69) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.098323 (rank : 70) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
PHF6_MOUSE
|
||||||
| NC score | 0.097807 (rank : 71) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.097754 (rank : 72) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.096927 (rank : 73) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
LBR_MOUSE
|
||||||
| NC score | 0.096357 (rank : 74) | θ value | 0.163984 (rank : 41) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3U9G9, Q3TSW2, Q811V8, Q811V9, Q8BST3, Q8K2Y8, Q8VDM0, Q91YS5, Q91Z27 | Gene names | Lbr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lamin-B receptor (Integral nuclear envelope inner membrane protein). | |||||
|
UHRF2_MOUSE
|
||||||
| NC score | 0.096269 (rank : 75) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.096050 (rank : 76) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.095841 (rank : 77) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.095083 (rank : 78) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.094358 (rank : 79) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.094175 (rank : 80) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.092534 (rank : 81) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.092345 (rank : 82) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
UHRF2_HUMAN
|
||||||
| NC score | 0.089238 (rank : 83) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.088989 (rank : 84) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.085444 (rank : 85) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.084851 (rank : 86) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.084809 (rank : 87) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.082935 (rank : 88) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
UHRF1_HUMAN
|
||||||
| NC score | 0.081525 (rank : 89) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
K1333_HUMAN
|
||||||
| NC score | 0.078553 (rank : 90) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.078247 (rank : 91) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.078061 (rank : 92) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.077650 (rank : 93) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
UHRF1_MOUSE
|
||||||
| NC score | 0.076673 (rank : 94) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.069294 (rank : 95) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
ARI5B_MOUSE
|
||||||
| NC score | 0.068208 (rank : 96) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.067627 (rank : 97) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.067043 (rank : 98) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.066407 (rank : 99) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
TIF1B_MOUSE
|
||||||
| NC score | 0.066090 (rank : 100) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
TIF1A_HUMAN
|
||||||
| NC score | 0.065517 (rank : 101) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.063561 (rank : 102) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1B_HUMAN
|
||||||
| NC score | 0.062082 (rank : 103) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.061914 (rank : 104) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.059850 (rank : 105) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.058226 (rank : 106) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
BRD9_HUMAN
|
||||||
| NC score | 0.057964 (rank : 107) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H8M2, Q2XUS1, Q6UWU9, Q8IUS4, Q9H5Q5, Q9H7R9 | Gene names | BRD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9 (Rhabdomyosarcoma antigen MU-RMS- 40.8). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.057473 (rank : 108) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
BRD7_HUMAN
|
||||||
| NC score | 0.056993 (rank : 109) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
PHF13_MOUSE
|
||||||
| NC score | 0.056253 (rank : 110) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K2W6 | Gene names | Phf13 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 13. | |||||
|
PHF13_HUMAN
|
||||||
| NC score | 0.056194 (rank : 111) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86YI8, Q8N551, Q9UJP2 | Gene names | PHF13 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 13. | |||||
|
BRD7_MOUSE
|
||||||
| NC score | 0.055566 (rank : 112) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88665, Q3UQ56, Q3UU06, Q9CT78 | Gene names | Brd7, Bp75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein). | |||||
|
BRD9_MOUSE
|
||||||
| NC score | 0.055420 (rank : 113) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UQU0, Q5FWH1, Q811F7 | Gene names | Brd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9. | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.053919 (rank : 114) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
EPC1_HUMAN
|
||||||
| NC score | 0.052013 (rank : 115) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H2F5, Q5VW54, Q5VW56, Q5VW58, Q8NAQ4, Q8NE21, Q96LF4, Q96RR6, Q9H7T7 | Gene names | EPC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 1. | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.048803 (rank : 116) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.041513 (rank : 117) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.038155 (rank : 118) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.034886 (rank : 119) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.033997 (rank : 120) | θ value | 0.0431538 (rank : 36) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.033685 (rank : 121) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
BSCL2_HUMAN
|
||||||
| NC score | 0.031474 (rank : 122) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
BSCL2_MOUSE
|
||||||
| NC score | 0.029743 (rank : 123) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2E9, Q810B0, Q9JJC2, Q9JMF1 | Gene names | Bscl2, Gng3lg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein homolog). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.029304 (rank : 124) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.029050 (rank : 125) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.028484 (rank : 126) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.028185 (rank : 127) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.026761 (rank : 128) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.025227 (rank : 129) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.024604 (rank : 130) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
CI040_MOUSE
|
||||||
| NC score | 0.024364 (rank : 131) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCE4, Q8BJJ5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf40 homolog. | |||||
|
WDR24_HUMAN
|
||||||
| NC score | 0.023483 (rank : 132) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96S15, Q96GC7, Q9H0B7 | Gene names | WDR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 24. | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.023051 (rank : 133) | θ value | 0.0148317 (rank : 35) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.023023 (rank : 134) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.022355 (rank : 135) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
TNAP2_HUMAN
|
||||||
| NC score | 0.020589 (rank : 136) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03169, Q86VI0 | Gene names | TNFAIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 2 (Primary response gene B94 protein). | |||||
|
RANB3_MOUSE
|
||||||
| NC score | 0.020020 (rank : 137) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CT10, Q3TIS4, Q3U2G4, Q3UYG7 | Gene names | Ranbp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.019812 (rank : 138) | θ value | 0.0113563 (rank : 33) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
LCE2A_HUMAN
|
||||||
| NC score | 0.019422 (rank : 139) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
FBX46_MOUSE
|
||||||
| NC score | 0.018940 (rank : 140) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
WDR24_MOUSE
|
||||||
| NC score | 0.018378 (rank : 141) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFJ9 | Gene names | Wdr24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 24. | |||||
|
PCGF6_HUMAN
|
||||||
| NC score | 0.017490 (rank : 142) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYE7, Q5SYD1, Q5SYD6, Q96ID9, Q96SJ1 | Gene names | PCGF6, MBLR, RNF134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb group RING finger protein 6 (RING finger protein 134) (Mel18 and Bmi1-like RING finger). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.015661 (rank : 143) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.014569 (rank : 144) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TF3B_HUMAN
|
||||||
| NC score | 0.014082 (rank : 145) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92994, Q13223, Q5PR24, Q6IQ02, Q9HCW6, Q9HCW7, Q9HCW8 | Gene names | BRF1, BRF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor IIIB 90 kDa subunit (TFIIIB90) (hTFIIIB90) (B- related factor 1) (BRF-1) (hBRF) (TATA box-binding protein-associated factor, RNA polymerase III, subunit 2) (TAF3B2). | |||||
|
RAVR1_MOUSE
|
||||||
| NC score | 0.013739 (rank : 146) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CW46, Q5DTT6, Q811K0, Q8C3Z1, Q8CA14 | Gene names | Raver1, Kiaa1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
TRM1_HUMAN
|
||||||
| NC score | 0.013684 (rank : 147) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXH9, O76103, Q548Y5, Q8WVA6 | Gene names | TRMT1 | |||
|
Domain Architecture |

|
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.013652 (rank : 148) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
APLP1_MOUSE
|
||||||
| NC score | 0.013650 (rank : 149) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
RAI2_HUMAN
|
||||||
| NC score | 0.013628 (rank : 150) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y5P3, Q8N6X7 | Gene names | RAI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.013219 (rank : 151) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.012901 (rank : 152) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
MUS81_MOUSE
|
||||||
| NC score | 0.012399 (rank : 153) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91ZJ0, Q8R356 | Gene names | Mus81 | |||
|
Domain Architecture |

|
|||||
| Description | Crossover junction endonuclease MUS81 (EC 3.1.22.-). | |||||
|
CCD45_MOUSE
|
||||||
| NC score | 0.011989 (rank : 154) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
TRM1_MOUSE
|
||||||
| NC score | 0.011877 (rank : 155) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TX08, Q3TBY4 | Gene names | Trmt1, D8Ertd812e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
PCGF6_MOUSE
|
||||||
| NC score | 0.010900 (rank : 156) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99NA9 | Gene names | Pcgf6, Mblr, Rnf134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb group RING finger protein 6 (RING finger protein 134) (Mel18 and Bmi1-like RING finger). | |||||
|
OTU7A_HUMAN
|
||||||
| NC score | 0.010393 (rank : 157) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.010096 (rank : 158) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.009638 (rank : 159) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
NCOA7_MOUSE
|
||||||
| NC score | 0.009513 (rank : 160) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6DFV7, Q3TAR1, Q3UNB3, Q66K05, Q6ZQM4, Q8BJ15, Q8BJ39 | Gene names | Ncoa7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7. | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.009206 (rank : 161) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
GNAS1_HUMAN
|
||||||
| NC score | 0.008010 (rank : 162) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5JWF2, O75684, O75685, Q5JW67, Q5JWF1, Q9NY42 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
TOPB1_MOUSE
|
||||||
| NC score | 0.007841 (rank : 163) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
PO6F1_MOUSE
|
||||||
| NC score | 0.007765 (rank : 164) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
JIP2_MOUSE
|
||||||
| NC score | 0.007629 (rank : 165) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
PTN22_MOUSE
|
||||||
| NC score | 0.007519 (rank : 166) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.007483 (rank : 167) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
PAIP1_HUMAN
|
||||||
| NC score | 0.007481 (rank : 168) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
DGCR8_HUMAN
|
||||||
| NC score | 0.007038 (rank : 169) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WYQ5, Q6DCB2, Q6MZE9, Q6Y2L0, Q96G39, Q96GP8, Q9H6L8, Q9H6T7, Q9NRW2 | Gene names | DGCR8, C22orf12, DGCRK6 | |||
|
Domain Architecture |

|
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.007031 (rank : 170) | θ value | 0.163984 (rank : 40) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.006767 (rank : 171) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
IL2RB_HUMAN
|
||||||
| NC score | 0.006739 (rank : 172) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14784 | Gene names | IL2RB | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-2 receptor beta chain precursor (IL-2 receptor) (P70-75) (p75) (High affinity IL-2 receptor subunit beta) (CD122 antigen). | |||||
|
RNZ2_HUMAN
|
||||||
| NC score | 0.006734 (rank : 173) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQ52, Q9HAS8, Q9HAS9, Q9NVT1 | Gene names | ELAC2, HPC2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc phosphodiesterase ELAC protein 2 (EC 3.1.26.11) (Ribonuclease Z 2) (RNase Z 2) (tRNase Z 2) (tRNA 3 endonuclease 2) (ElaC homolog protein 2) (Heredity prostate cancer protein 2). | |||||
|
FARP2_MOUSE
|
||||||
| NC score | 0.006207 (rank : 174) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.005908 (rank : 175) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
WDR46_HUMAN
|
||||||
| NC score | 0.005824 (rank : 176) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15213, Q5STK5 | Gene names | WDR46, BING4, C6orf11 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.004808 (rank : 177) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.002753 (rank : 178) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
INSM1_HUMAN
|
||||||
| NC score | 0.002274 (rank : 179) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 683 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q01101 | Gene names | INSM1, IA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.001504 (rank : 180) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
ZN447_HUMAN
|
||||||
| NC score | -0.001850 (rank : 181) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
KLF13_MOUSE
|
||||||
| NC score | -0.002553 (rank : 182) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJZ6, Q9ESX3, Q9JHF8 | Gene names | Klf13, Bteb3, Fklf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 13 (Transcription factor BTEB3) (Basic transcription element-binding protein 3) (BTE-binding protein 3) (RANTES factor of late activated T-lymphocytes 1) (RFLAT-1) (Erythroid transcription factor FKLF-2). | |||||
|
PKN3_MOUSE
|
||||||
| NC score | -0.003069 (rank : 183) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
HKR3_HUMAN
|
||||||
| NC score | -0.003211 (rank : 184) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10074 | Gene names | HKR3 | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-related zinc finger protein 3 (Protein HKR3). | |||||