Please be patient as the page loads
|
DGCR8_HUMAN
|
||||||
| SwissProt Accessions | Q8WYQ5, Q6DCB2, Q6MZE9, Q6Y2L0, Q96G39, Q96GP8, Q9H6L8, Q9H6T7, Q9NRW2 | Gene names | DGCR8, C22orf12, DGCRK6 | |||
|
Domain Architecture |

|
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DGCR8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8WYQ5, Q6DCB2, Q6MZE9, Q6Y2L0, Q96G39, Q96GP8, Q9H6L8, Q9H6T7, Q9NRW2 | Gene names | DGCR8, C22orf12, DGCRK6 | |||
|
Domain Architecture |

|
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8). | |||||
|
DGCR8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990863 (rank : 2) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9EQM6 | Gene names | Dgcr8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8 homolog) (Gy1). | |||||
|
PRKRA_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 3) | NC score | 0.178965 (rank : 3) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75569, Q53G24, Q6X7T5, Q8NDK4 | Gene names | PRKRA, PACT, RAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X). | |||||
|
PRKRA_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 4) | NC score | 0.178647 (rank : 4) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WTX2, Q9CZB7 | Gene names | Prkra, Rax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X) (RAX). | |||||
|
LRRF1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 5) | NC score | 0.100061 (rank : 9) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
DSRAD_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 6) | NC score | 0.091282 (rank : 13) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
|
WWP1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.093082 (rank : 12) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
AN32A_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.103064 (rank : 7) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35381, P97437 | Gene names | Anp32a, Anp32, Lanp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein). | |||||
|
AN32C_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.103064 (rank : 8) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64G17 | Gene names | Anp32c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C. | |||||
|
HCLS1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.050996 (rank : 37) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P49710 | Gene names | Hcls1, Hs1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1). | |||||
|
NED4L_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.078959 (rank : 18) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
AN32B_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.097137 (rank : 10) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
ITCH_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.087550 (rank : 15) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
ITCH_MOUSE
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.088213 (rank : 14) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.087184 (rank : 16) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.072027 (rank : 21) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.023226 (rank : 54) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ERBB2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.009505 (rank : 67) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70424, Q61525, Q6ZPE0 | Gene names | Erbb2, Kiaa3023, Neu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.022373 (rank : 55) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
WWTR1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.110281 (rank : 6) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9GZV5, Q8N3P2, Q9Y3W6 | Gene names | WWTR1, TAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
WWTR1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.111621 (rank : 5) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
CABL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.058096 (rank : 31) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.069238 (rank : 24) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
K1914_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.041367 (rank : 40) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N4X5, Q8TB54, Q96PX4, Q96SY5 | Gene names | KIAA1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
NED4L_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.072896 (rank : 20) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.010784 (rank : 66) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
AN32A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.078985 (rank : 17) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39687 | Gene names | ANP32A, C15orf1, LANP, MAPM, PHAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein) (Lanp) (Putative HLA-DR-associated protein I) (PHAPI) (Mapmodulin). | |||||
|
C2TA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.041266 (rank : 41) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
OFD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.019358 (rank : 58) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.070586 (rank : 23) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
NEDD4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.070825 (rank : 22) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.014076 (rank : 64) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
LMO7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.037855 (rank : 42) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
NRIP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.035850 (rank : 44) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CBD1, Q8C3Y8, Q9Z2K2 | Gene names | Nrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.041596 (rank : 39) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.019681 (rank : 56) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
YAP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.093202 (rank : 11) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
ZN710_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.004323 (rank : 80) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N1W2, Q6ZMK9, Q8NDU0 | Gene names | ZNF710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.006088 (rank : 75) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
EGR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.004541 (rank : 78) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11161, Q8IV26, Q9UNA6 | Gene names | EGR2, KROX20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein) (AT591). | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.007231 (rank : 71) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
EGR2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.004473 (rank : 79) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08152 | Gene names | Egr2, Egr-2, Krox-20, Zfp-25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein). | |||||
|
GMIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.011895 (rank : 65) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GP101_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.006576 (rank : 74) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80T62, Q3UUI9 | Gene names | Gpr101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 101. | |||||
|
HCLS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.035635 (rank : 45) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14317 | Gene names | HCLS1, HS1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1) (p75). | |||||
|
HS90A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.019523 (rank : 57) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.037119 (rank : 43) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
ZN365_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.018647 (rank : 59) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q70YC5, O94930, Q5HYE6, Q68SG8, Q6NSK2, Q6P9D4, Q70YC6, Q70YC7 | Gene names | ZNF365, KIAA0844 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365. | |||||
|
ZN710_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.003606 (rank : 81) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3U288, Q3U341, Q6P9R3, Q8BJH0, Q8BTL6 | Gene names | Znf710, Zfp710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
AN32B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.064550 (rank : 25) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
CABL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.031349 (rank : 51) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESJ1, Q9EPR8 | Gene names | Cables1, Cables | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
EDA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.014608 (rank : 62) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
K0310_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.030728 (rank : 52) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
K1914_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.032768 (rank : 48) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5DTU0, Q8BID1, Q8K2G0 | Gene names | Kiaa1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
KI13B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.005746 (rank : 76) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
LMD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.015229 (rank : 61) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29536 | Gene names | LMOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Leiomodin-1 (Leiomodin, muscle form) (64 kDa autoantigen D1) (64 kDa autoantigen 1D) (64 kDa autoantigen 1D3) (Thyroid-associated ophthalmopathy autoantigen) (Smooth muscle leiomodin) (SM-Lmod). | |||||
|
NRIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.030665 (rank : 53) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48552, Q8IWE8 | Gene names | NRIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
PAF49_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.031901 (rank : 50) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
U2AFM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.014377 (rank : 63) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
YAP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.078544 (rank : 19) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
ZN408_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.001490 (rank : 82) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H9D4 | Gene names | ZNF408, PFM14, PRDM17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 408 (PR-domain zinc finger protein 17). | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.007593 (rank : 70) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
JHD3B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.007038 (rank : 72) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.006647 (rank : 73) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.007648 (rank : 69) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
PDE4C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.004887 (rank : 77) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
PQBP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.015274 (rank : 60) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VJ5, Q80WW2, Q9ER43, Q9QYY2 | Gene names | Pqbp1, Npw38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38). | |||||
|
PTMA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.032158 (rank : 49) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
SRC8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.033561 (rank : 47) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14247 | Gene names | CTTN, EMS1 | |||
|
Domain Architecture |
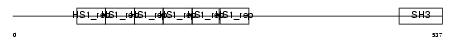
|
|||||
| Description | Src substrate cortactin (Amplaxin) (Oncogene EMS1). | |||||
|
SRC8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.033600 (rank : 46) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60598 | Gene names | Cttn, Ems1 | |||
|
Domain Architecture |
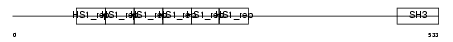
|
|||||
| Description | Src substrate cortactin. | |||||
|
STAU1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.041940 (rank : 38) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.009339 (rank : 68) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
AN32C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.054443 (rank : 35) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43423 | Gene names | ANP32C, PP32R1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C (Phosphoprotein 32-related protein 1) (Tumorigenic protein pp32r1). | |||||
|
AN32D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.054318 (rank : 36) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95626, Q6NTC4 | Gene names | ANP32D, PP32R2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member D (Phosphoprotein 32-related protein 2) (Tumorigenic protein pp32r2). | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.058769 (rank : 30) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
SMUF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.056711 (rank : 34) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.056715 (rank : 33) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
SMUF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.057470 (rank : 32) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
TRBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.063462 (rank : 27) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15633, Q12878 | Gene names | TARBP2, TRBP | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Trans-activation-responsive RNA-binding protein). | |||||
|
TRBP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.063028 (rank : 29) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97473 | Gene names | Tarbp2, Prbp | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Protamine-1 RNA-binding protein) (PRM-1 RNA-binding protein). | |||||
|
WWP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.063853 (rank : 26) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
WWP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.063374 (rank : 28) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
DGCR8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8WYQ5, Q6DCB2, Q6MZE9, Q6Y2L0, Q96G39, Q96GP8, Q9H6L8, Q9H6T7, Q9NRW2 | Gene names | DGCR8, C22orf12, DGCRK6 | |||
|
Domain Architecture |

|
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8). | |||||
|
DGCR8_MOUSE
|
||||||
| NC score | 0.990863 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9EQM6 | Gene names | Dgcr8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8 homolog) (Gy1). | |||||
|
PRKRA_HUMAN
|
||||||
| NC score | 0.178965 (rank : 3) | θ value | 0.00134147 (rank : 3) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75569, Q53G24, Q6X7T5, Q8NDK4 | Gene names | PRKRA, PACT, RAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X). | |||||
|
PRKRA_MOUSE
|
||||||
| NC score | 0.178647 (rank : 4) | θ value | 0.00134147 (rank : 4) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WTX2, Q9CZB7 | Gene names | Prkra, Rax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X) (RAX). | |||||
|
WWTR1_MOUSE
|
||||||
| NC score | 0.111621 (rank : 5) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
WWTR1_HUMAN
|
||||||
| NC score | 0.110281 (rank : 6) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9GZV5, Q8N3P2, Q9Y3W6 | Gene names | WWTR1, TAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
AN32A_MOUSE
|
||||||
| NC score | 0.103064 (rank : 7) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35381, P97437 | Gene names | Anp32a, Anp32, Lanp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein). | |||||
|
AN32C_MOUSE
|
||||||
| NC score | 0.103064 (rank : 8) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64G17 | Gene names | Anp32c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C. | |||||
|
LRRF1_HUMAN
|
||||||
| NC score | 0.100061 (rank : 9) | θ value | 0.00869519 (rank : 5) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
AN32B_HUMAN
|
||||||
| NC score | 0.097137 (rank : 10) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
YAP1_MOUSE
|
||||||
| NC score | 0.093202 (rank : 11) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
WWP1_MOUSE
|
||||||
| NC score | 0.093082 (rank : 12) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
DSRAD_HUMAN
|
||||||
| NC score | 0.091282 (rank : 13) | θ value | 0.0431538 (rank : 6) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
|
ITCH_MOUSE
|
||||||
| NC score | 0.088213 (rank : 14) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
ITCH_HUMAN
|
||||||
| NC score | 0.087550 (rank : 15) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.087184 (rank : 16) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
AN32A_HUMAN
|
||||||
| NC score | 0.078985 (rank : 17) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39687 | Gene names | ANP32A, C15orf1, LANP, MAPM, PHAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein) (Lanp) (Putative HLA-DR-associated protein I) (PHAPI) (Mapmodulin). | |||||
|
NED4L_HUMAN
|
||||||
| NC score | 0.078959 (rank : 18) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
YAP1_HUMAN
|
||||||
| NC score | 0.078544 (rank : 19) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
NED4L_MOUSE
|
||||||
| NC score | 0.072896 (rank : 20) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.072027 (rank : 21) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
NEDD4_MOUSE
|
||||||
| NC score | 0.070825 (rank : 22) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.070586 (rank : 23) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.069238 (rank : 24) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
AN32B_MOUSE
|
||||||
| NC score | 0.064550 (rank : 25) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.063853 (rank : 26) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
TRBP2_HUMAN
|
||||||
| NC score | 0.063462 (rank : 27) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15633, Q12878 | Gene names | TARBP2, TRBP | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Trans-activation-responsive RNA-binding protein). | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.063374 (rank : 28) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
TRBP2_MOUSE
|
||||||
| NC score | 0.063028 (rank : 29) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97473 | Gene names | Tarbp2, Prbp | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Protamine-1 RNA-binding protein) (PRM-1 RNA-binding protein). | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.058769 (rank : 30) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
CABL1_HUMAN
|
||||||
| NC score | 0.058096 (rank : 31) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
SMUF2_HUMAN
|
||||||
| NC score | 0.057470 (rank : 32) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
SMUF1_MOUSE
|
||||||
| NC score | 0.056715 (rank : 33) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
SMUF1_HUMAN
|
||||||
| NC score | 0.056711 (rank : 34) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
AN32C_HUMAN
|
||||||
| NC score | 0.054443 (rank : 35) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43423 | Gene names | ANP32C, PP32R1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C (Phosphoprotein 32-related protein 1) (Tumorigenic protein pp32r1). | |||||
|
AN32D_HUMAN
|
||||||
| NC score | 0.054318 (rank : 36) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95626, Q6NTC4 | Gene names | ANP32D, PP32R2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member D (Phosphoprotein 32-related protein 2) (Tumorigenic protein pp32r2). | |||||
|
HCLS1_MOUSE
|
||||||
| NC score | 0.050996 (rank : 37) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P49710 | Gene names | Hcls1, Hs1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1). | |||||
|
STAU1_HUMAN
|
||||||
| NC score | 0.041940 (rank : 38) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.041596 (rank : 39) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
K1914_HUMAN
|
||||||
| NC score | 0.041367 (rank : 40) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N4X5, Q8TB54, Q96PX4, Q96SY5 | Gene names | KIAA1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
C2TA_MOUSE
|
||||||
| NC score | 0.041266 (rank : 41) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
LMO7_HUMAN
|
||||||
| NC score | 0.037855 (rank : 42) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.037119 (rank : 43) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
NRIP1_MOUSE
|
||||||
| NC score | 0.035850 (rank : 44) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CBD1, Q8C3Y8, Q9Z2K2 | Gene names | Nrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
HCLS1_HUMAN
|
||||||
| NC score | 0.035635 (rank : 45) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14317 | Gene names | HCLS1, HS1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1) (p75). | |||||
|
SRC8_MOUSE
|
||||||
| NC score | 0.033600 (rank : 46) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60598 | Gene names | Cttn, Ems1 | |||
|
Domain Architecture |
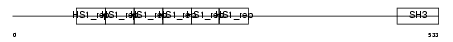
|
|||||
| Description | Src substrate cortactin. | |||||
|
SRC8_HUMAN
|
||||||
| NC score | 0.033561 (rank : 47) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14247 | Gene names | CTTN, EMS1 | |||
|
Domain Architecture |
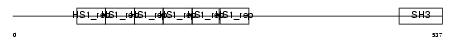
|
|||||
| Description | Src substrate cortactin (Amplaxin) (Oncogene EMS1). | |||||
|
K1914_MOUSE
|
||||||
| NC score | 0.032768 (rank : 48) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5DTU0, Q8BID1, Q8K2G0 | Gene names | Kiaa1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.032158 (rank : 49) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PAF49_HUMAN
|
||||||
| NC score | 0.031901 (rank : 50) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
CABL1_MOUSE
|
||||||
| NC score | 0.031349 (rank : 51) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESJ1, Q9EPR8 | Gene names | Cables1, Cables | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.030728 (rank : 52) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
NRIP1_HUMAN
|
||||||
| NC score | 0.030665 (rank : 53) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48552, Q8IWE8 | Gene names | NRIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.023226 (rank : 54) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.022373 (rank : 55) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.019681 (rank : 56) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
HS90A_MOUSE
|
||||||
| NC score | 0.019523 (rank : 57) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
|
OFD1_HUMAN
|
||||||
| NC score | 0.019358 (rank : 58) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
ZN365_HUMAN
|
||||||
| NC score | 0.018647 (rank : 59) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q70YC5, O94930, Q5HYE6, Q68SG8, Q6NSK2, Q6P9D4, Q70YC6, Q70YC7 | Gene names | ZNF365, KIAA0844 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365. | |||||
|
PQBP1_MOUSE
|
||||||
| NC score | 0.015274 (rank : 60) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VJ5, Q80WW2, Q9ER43, Q9QYY2 | Gene names | Pqbp1, Npw38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38). | |||||
|
LMD1_HUMAN
|
||||||
| NC score | 0.015229 (rank : 61) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29536 | Gene names | LMOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Leiomodin-1 (Leiomodin, muscle form) (64 kDa autoantigen D1) (64 kDa autoantigen 1D) (64 kDa autoantigen 1D3) (Thyroid-associated ophthalmopathy autoantigen) (Smooth muscle leiomodin) (SM-Lmod). | |||||
|
EDA_MOUSE
|
||||||
| NC score | 0.014608 (rank : 62) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
U2AFM_MOUSE
|
||||||
| NC score | 0.014377 (rank : 63) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.014076 (rank : 64) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
GMIP_HUMAN
|
||||||
| NC score | 0.011895 (rank : 65) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.010784 (rank : 66) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ERBB2_MOUSE
|
||||||
| NC score | 0.009505 (rank : 67) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70424, Q61525, Q6ZPE0 | Gene names | Erbb2, Kiaa3023, Neu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene). | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.009339 (rank : 68) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.007648 (rank : 69) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.007593 (rank : 70) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | 0.007231 (rank : 71) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
JHD3B_HUMAN
|
||||||
| NC score | 0.007038 (rank : 72) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.006647 (rank : 73) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
GP101_MOUSE
|
||||||
| NC score | 0.006576 (rank : 74) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80T62, Q3UUI9 | Gene names | Gpr101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 101. | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.006088 (rank : 75) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
KI13B_HUMAN
|
||||||
| NC score | 0.005746 (rank : 76) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
PDE4C_HUMAN
|
||||||
| NC score | 0.004887 (rank : 77) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
EGR2_HUMAN
|
||||||
| NC score | 0.004541 (rank : 78) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11161, Q8IV26, Q9UNA6 | Gene names | EGR2, KROX20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein) (AT591). | |||||
|
EGR2_MOUSE
|
||||||
| NC score | 0.004473 (rank : 79) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08152 | Gene names | Egr2, Egr-2, Krox-20, Zfp-25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein). | |||||
|
ZN710_HUMAN
|
||||||
| NC score | 0.004323 (rank : 80) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N1W2, Q6ZMK9, Q8NDU0 | Gene names | ZNF710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
ZN710_MOUSE
|
||||||
| NC score | 0.003606 (rank : 81) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3U288, Q3U341, Q6P9R3, Q8BJH0, Q8BTL6 | Gene names | Znf710, Zfp710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
ZN408_HUMAN
|
||||||
| NC score | 0.001490 (rank : 82) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H9D4 | Gene names | ZNF408, PFM14, PRDM17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 408 (PR-domain zinc finger protein 17). | |||||