Please be patient as the page loads
|
DSRAD_HUMAN
|
||||||
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DSRAD_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
|
DSRAD_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.982060 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
RED1_MOUSE
|
||||||
| θ value | 1.00825e-67 (rank : 3) | NC score | 0.842149 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91ZS8, Q8K3X1, Q91ZS6, Q91ZS7, Q91ZS9, Q99MU8 | Gene names | Adarb1, Adar2, Red1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
RED2_HUMAN
|
||||||
| θ value | 3.24342e-66 (rank : 4) | NC score | 0.845223 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NS39 | Gene names | ADARB2, ADAR3, RED2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
RED1_HUMAN
|
||||||
| θ value | 1.6097e-65 (rank : 5) | NC score | 0.838066 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78563, O00395, O00465, O00691, O00692, P78555, Q8NFD1 | Gene names | ADARB1, ADAR2, DRADA2, RED1 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
RED2_MOUSE
|
||||||
| θ value | 2.84138e-62 (rank : 6) | NC score | 0.835816 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JI20, Q3UTP1, Q8CC51, Q9JIE5 | Gene names | Adarb2, Adar3, Red2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
E2AK2_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 7) | NC score | 0.058813 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19525 | Gene names | EIF2AK2, PKR, PRKR | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase). | |||||
|
E2AK2_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 8) | NC score | 0.043539 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03963, Q61742, Q62026 | Gene names | Eif2ak2, Pkr, Prkr, Tik | |||
|
Domain Architecture |
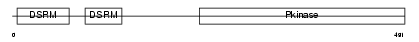
|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase) (Serine/threonine-protein kinase TIK). | |||||
|
PRKRA_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 9) | NC score | 0.361560 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WTX2, Q9CZB7 | Gene names | Prkra, Rax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X) (RAX). | |||||
|
STAU2_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 10) | NC score | 0.405489 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CJ67, Q8BSY8, Q8CJ66, Q8R175, Q91Z19, Q9D5N7 | Gene names | Stau2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
PRKRA_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 11) | NC score | 0.360485 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75569, Q53G24, Q6X7T5, Q8NDK4 | Gene names | PRKRA, PACT, RAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X). | |||||
|
ILF3_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 12) | NC score | 0.459685 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
ILF3_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 13) | NC score | 0.459701 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
TRBP2_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 14) | NC score | 0.352422 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97473 | Gene names | Tarbp2, Prbp | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Protamine-1 RNA-binding protein) (PRM-1 RNA-binding protein). | |||||
|
TRBP2_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 15) | NC score | 0.335001 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15633, Q12878 | Gene names | TARBP2, TRBP | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Trans-activation-responsive RNA-binding protein). | |||||
|
STAU2_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 16) | NC score | 0.380507 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NUL3, Q6AHY7, Q96HM0, Q96HM1, Q9NVI5, Q9UGG6 | Gene names | STAU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
STAU1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 17) | NC score | 0.271868 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z108 | Gene names | Stau1, Stau | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
STAU1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 18) | NC score | 0.276470 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
CM003_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 19) | NC score | 0.070945 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IX90, Q5VZV5, Q86WR2, Q8NBG1, Q96D22 | Gene names | C13orf3, RAMA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf3. | |||||
|
DGCR8_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 20) | NC score | 0.091282 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WYQ5, Q6DCB2, Q6MZE9, Q6Y2L0, Q96G39, Q96GP8, Q9H6L8, Q9H6T7, Q9NRW2 | Gene names | DGCR8, C22orf12, DGCRK6 | |||
|
Domain Architecture |

|
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8). | |||||
|
DGCR8_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.091339 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQM6 | Gene names | Dgcr8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8 homolog) (Gy1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.062723 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
ZBP1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.144528 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QY24, Q8VE02, Q9D8B9 | Gene names | Zbp1, Dlm1 | |||
|
Domain Architecture |
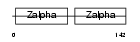
|
|||||
| Description | Z-DNA-binding protein 1 (Tumor stroma and activated macrophage protein DLM-1). | |||||
|
TGON1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.033695 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
CQ028_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.033066 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R1F6, Q8C8V6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 homolog. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.022055 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PEX14_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.049621 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75381, Q8WX51 | Gene names | PEX14 | |||
|
Domain Architecture |
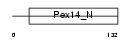
|
|||||
| Description | Peroxisomal membrane protein PEX14 (Peroxin-14) (Peroxisomal membrane anchor protein PEX14) (PTS1 receptor docking protein). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.035830 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
DGKG_MOUSE
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.014793 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WG7 | Gene names | Dgkg, Dagk3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma) (88 kDa diacylglycerol kinase). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.015331 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.010957 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NKRF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.060667 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
CRSP6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.032786 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NVC6, Q9UNP7, Q9Y2W0, Q9Y660 | Gene names | CRSP6, ARC77, DRIP77, DRIP80, TRAP80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 6 (Cofactor required for Sp1 transcriptional activation subunit 6) (Transcriptional coactivator CRSP77) (Vitamin D3 receptor-interacting protein complex 80 kDa component) (DRIP80) (Thyroid hormone receptor-associated protein complex 80 kDa component) (Trap80) (Activator-recruited cofactor 77 kDa component) (ARC77). | |||||
|
FIBA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.009681 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P02671, Q9BX62, Q9UCH2 | Gene names | FGA | |||
|
Domain Architecture |

|
|||||
| Description | Fibrinogen alpha chain precursor [Contains: Fibrinopeptide A]. | |||||
|
LRP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.005088 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
PAX9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.013776 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47242 | Gene names | Pax9, Pax-9 | |||
|
Domain Architecture |
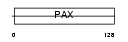
|
|||||
| Description | Paired box protein Pax-9. | |||||
|
PEX14_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.038963 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0A0 | Gene names | Pex14 | |||
|
Domain Architecture |
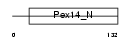
|
|||||
| Description | Peroxisomal membrane protein PEX14 (Peroxin-14) (Peroxisomal membrane anchor protein PEX14) (PTS1 receptor docking protein). | |||||
|
ZBP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.088935 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H171, Q5JY39, Q9BYW4 | Gene names | ZBP1, C20orf183, DLM1 | |||
|
Domain Architecture |
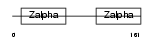
|
|||||
| Description | Z-DNA-binding protein 1 (Tumor stroma and activated macrophage protein DLM-1). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.002597 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.012859 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
PHAR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.017365 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.009878 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SEPT9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.014742 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
IMA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.010382 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |
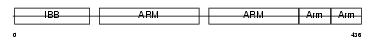
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
LACTB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.016390 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P83111, P83096 | Gene names | LACTB | |||
|
Domain Architecture |
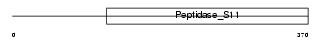
|
|||||
| Description | Serine beta-lactamase-like protein LACTB. | |||||
|
M10L1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.010609 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXT6, Q5TGD5, Q8NBD4, Q9NXW3, Q9UFB3, Q9UGX9 | Gene names | MOV10L1 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase Mov10l1 (EC 3.6.1.-) (Moloney leukemia virus 10-like protein 1) (MOV10-like 1). | |||||
|
AP180_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.012915 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.006240 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
NFAC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.008179 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
PAX9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.011381 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55771, Q99582, Q9UQR4 | Gene names | PAX9 | |||
|
Domain Architecture |
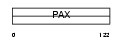
|
|||||
| Description | Paired box protein Pax-9. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.006804 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PGCB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.004627 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
RHBT3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.008822 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CTN4, Q80X55, Q9CVT0 | Gene names | Rhobtb3, Kiaa0878 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-related BTB domain-containing protein 3. | |||||
|
SART3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.006920 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15020, Q58F06, Q8IUS1, Q96J95 | Gene names | SART3, KIAA0156, TIP110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (hSART-3) (Tat-interacting protein of 110 kDa) (Tip110). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.009807 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
HMCN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | -0.000254 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||
|
IMA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.008899 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |
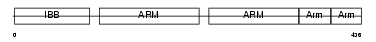
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
NKRF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.031722 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
DSRAD_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
|
DSRAD_MOUSE
|
||||||
| NC score | 0.982060 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
RED2_HUMAN
|
||||||
| NC score | 0.845223 (rank : 3) | θ value | 3.24342e-66 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NS39 | Gene names | ADARB2, ADAR3, RED2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
RED1_MOUSE
|
||||||
| NC score | 0.842149 (rank : 4) | θ value | 1.00825e-67 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91ZS8, Q8K3X1, Q91ZS6, Q91ZS7, Q91ZS9, Q99MU8 | Gene names | Adarb1, Adar2, Red1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
RED1_HUMAN
|
||||||
| NC score | 0.838066 (rank : 5) | θ value | 1.6097e-65 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78563, O00395, O00465, O00691, O00692, P78555, Q8NFD1 | Gene names | ADARB1, ADAR2, DRADA2, RED1 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
RED2_MOUSE
|
||||||
| NC score | 0.835816 (rank : 6) | θ value | 2.84138e-62 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JI20, Q3UTP1, Q8CC51, Q9JIE5 | Gene names | Adarb2, Adar3, Red2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
ILF3_HUMAN
|
||||||
| NC score | 0.459701 (rank : 7) | θ value | 2.44474e-05 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
ILF3_MOUSE
|
||||||
| NC score | 0.459685 (rank : 8) | θ value | 6.43352e-06 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
STAU2_MOUSE
|
||||||
| NC score | 0.405489 (rank : 9) | θ value | 9.92553e-07 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CJ67, Q8BSY8, Q8CJ66, Q8R175, Q91Z19, Q9D5N7 | Gene names | Stau2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
STAU2_HUMAN
|
||||||
| NC score | 0.380507 (rank : 10) | θ value | 0.000158464 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NUL3, Q6AHY7, Q96HM0, Q96HM1, Q9NVI5, Q9UGG6 | Gene names | STAU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
PRKRA_MOUSE
|
||||||
| NC score | 0.361560 (rank : 11) | θ value | 9.92553e-07 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WTX2, Q9CZB7 | Gene names | Prkra, Rax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X) (RAX). | |||||
|
PRKRA_HUMAN
|
||||||
| NC score | 0.360485 (rank : 12) | θ value | 1.29631e-06 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75569, Q53G24, Q6X7T5, Q8NDK4 | Gene names | PRKRA, PACT, RAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X). | |||||
|
TRBP2_MOUSE
|
||||||
| NC score | 0.352422 (rank : 13) | θ value | 2.44474e-05 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97473 | Gene names | Tarbp2, Prbp | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Protamine-1 RNA-binding protein) (PRM-1 RNA-binding protein). | |||||
|
TRBP2_HUMAN
|
||||||
| NC score | 0.335001 (rank : 14) | θ value | 4.1701e-05 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15633, Q12878 | Gene names | TARBP2, TRBP | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Trans-activation-responsive RNA-binding protein). | |||||
|
STAU1_HUMAN
|
||||||
| NC score | 0.276470 (rank : 15) | θ value | 0.00869519 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
STAU1_MOUSE
|
||||||
| NC score | 0.271868 (rank : 16) | θ value | 0.00509761 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z108 | Gene names | Stau1, Stau | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
ZBP1_MOUSE
|
||||||
| NC score | 0.144528 (rank : 17) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QY24, Q8VE02, Q9D8B9 | Gene names | Zbp1, Dlm1 | |||
|
Domain Architecture |

|
|||||
| Description | Z-DNA-binding protein 1 (Tumor stroma and activated macrophage protein DLM-1). | |||||
|
DGCR8_MOUSE
|
||||||
| NC score | 0.091339 (rank : 18) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQM6 | Gene names | Dgcr8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8 homolog) (Gy1). | |||||
|
DGCR8_HUMAN
|
||||||
| NC score | 0.091282 (rank : 19) | θ value | 0.0431538 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WYQ5, Q6DCB2, Q6MZE9, Q6Y2L0, Q96G39, Q96GP8, Q9H6L8, Q9H6T7, Q9NRW2 | Gene names | DGCR8, C22orf12, DGCRK6 | |||
|
Domain Architecture |

|
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8). | |||||
|
ZBP1_HUMAN
|
||||||
| NC score | 0.088935 (rank : 20) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H171, Q5JY39, Q9BYW4 | Gene names | ZBP1, C20orf183, DLM1 | |||
|
Domain Architecture |

|
|||||
| Description | Z-DNA-binding protein 1 (Tumor stroma and activated macrophage protein DLM-1). | |||||
|
CM003_HUMAN
|
||||||
| NC score | 0.070945 (rank : 21) | θ value | 0.0193708 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IX90, Q5VZV5, Q86WR2, Q8NBG1, Q96D22 | Gene names | C13orf3, RAMA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf3. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.062723 (rank : 22) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
NKRF_HUMAN
|
||||||
| NC score | 0.060667 (rank : 23) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
E2AK2_HUMAN
|
||||||
| NC score | 0.058813 (rank : 24) | θ value | 1.25267e-09 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19525 | Gene names | EIF2AK2, PKR, PRKR | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase). | |||||
|
PEX14_HUMAN
|
||||||
| NC score | 0.049621 (rank : 25) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75381, Q8WX51 | Gene names | PEX14 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal membrane protein PEX14 (Peroxin-14) (Peroxisomal membrane anchor protein PEX14) (PTS1 receptor docking protein). | |||||
|
E2AK2_MOUSE
|
||||||
| NC score | 0.043539 (rank : 26) | θ value | 9.92553e-07 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03963, Q61742, Q62026 | Gene names | Eif2ak2, Pkr, Prkr, Tik | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase) (Serine/threonine-protein kinase TIK). | |||||
|
PEX14_MOUSE
|
||||||
| NC score | 0.038963 (rank : 27) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0A0 | Gene names | Pex14 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal membrane protein PEX14 (Peroxin-14) (Peroxisomal membrane anchor protein PEX14) (PTS1 receptor docking protein). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.035830 (rank : 28) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.033695 (rank : 29) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
CQ028_MOUSE
|
||||||
| NC score | 0.033066 (rank : 30) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R1F6, Q8C8V6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 homolog. | |||||
|
CRSP6_HUMAN
|
||||||
| NC score | 0.032786 (rank : 31) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NVC6, Q9UNP7, Q9Y2W0, Q9Y660 | Gene names | CRSP6, ARC77, DRIP77, DRIP80, TRAP80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 6 (Cofactor required for Sp1 transcriptional activation subunit 6) (Transcriptional coactivator CRSP77) (Vitamin D3 receptor-interacting protein complex 80 kDa component) (DRIP80) (Thyroid hormone receptor-associated protein complex 80 kDa component) (Trap80) (Activator-recruited cofactor 77 kDa component) (ARC77). | |||||
|
NKRF_MOUSE
|
||||||
| NC score | 0.031722 (rank : 32) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.022055 (rank : 33) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PHAR2_HUMAN
|
||||||
| NC score | 0.017365 (rank : 34) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
LACTB_HUMAN
|
||||||
| NC score | 0.016390 (rank : 35) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P83111, P83096 | Gene names | LACTB | |||
|
Domain Architecture |

|
|||||
| Description | Serine beta-lactamase-like protein LACTB. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.015331 (rank : 36) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
DGKG_MOUSE
|
||||||
| NC score | 0.014793 (rank : 37) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WG7 | Gene names | Dgkg, Dagk3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma) (88 kDa diacylglycerol kinase). | |||||
|
SEPT9_MOUSE
|
||||||
| NC score | 0.014742 (rank : 38) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
PAX9_MOUSE
|
||||||
| NC score | 0.013776 (rank : 39) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47242 | Gene names | Pax9, Pax-9 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-9. | |||||
|
AP180_HUMAN
|
||||||
| NC score | 0.012915 (rank : 40) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.012859 (rank : 41) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
PAX9_HUMAN
|
||||||
| NC score | 0.011381 (rank : 42) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55771, Q99582, Q9UQR4 | Gene names | PAX9 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-9. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.010957 (rank : 43) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
M10L1_HUMAN
|
||||||
| NC score | 0.010609 (rank : 44) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXT6, Q5TGD5, Q8NBD4, Q9NXW3, Q9UFB3, Q9UGX9 | Gene names | MOV10L1 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase Mov10l1 (EC 3.6.1.-) (Moloney leukemia virus 10-like protein 1) (MOV10-like 1). | |||||
|
IMA2_MOUSE
|
||||||
| NC score | 0.010382 (rank : 45) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.009878 (rank : 46) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.009807 (rank : 47) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
FIBA_HUMAN
|
||||||
| NC score | 0.009681 (rank : 48) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P02671, Q9BX62, Q9UCH2 | Gene names | FGA | |||
|
Domain Architecture |

|
|||||
| Description | Fibrinogen alpha chain precursor [Contains: Fibrinopeptide A]. | |||||
|
IMA2_HUMAN
|
||||||
| NC score | 0.008899 (rank : 49) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
RHBT3_MOUSE
|
||||||
| NC score | 0.008822 (rank : 50) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CTN4, Q80X55, Q9CVT0 | Gene names | Rhobtb3, Kiaa0878 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-related BTB domain-containing protein 3. | |||||
|
NFAC2_HUMAN
|
||||||
| NC score | 0.008179 (rank : 51) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
SART3_HUMAN
|
||||||
| NC score | 0.006920 (rank : 52) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15020, Q58F06, Q8IUS1, Q96J95 | Gene names | SART3, KIAA0156, TIP110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (hSART-3) (Tat-interacting protein of 110 kDa) (Tip110). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.006804 (rank : 53) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.006240 (rank : 54) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.005088 (rank : 55) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
PGCB_HUMAN
|
||||||
| NC score | 0.004627 (rank : 56) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.002597 (rank : 57) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
HMCN1_HUMAN
|
||||||
| NC score | -0.000254 (rank : 58) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||