Please be patient as the page loads
|
RED1_MOUSE
|
||||||
| SwissProt Accessions | Q91ZS8, Q8K3X1, Q91ZS6, Q91ZS7, Q91ZS9, Q99MU8 | Gene names | Adarb1, Adar2, Red1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RED1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994252 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P78563, O00395, O00465, O00691, O00692, P78555, Q8NFD1 | Gene names | ADARB1, ADAR2, DRADA2, RED1 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
RED1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91ZS8, Q8K3X1, Q91ZS6, Q91ZS7, Q91ZS9, Q99MU8 | Gene names | Adarb1, Adar2, Red1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
RED2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.982121 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NS39 | Gene names | ADARB2, ADAR3, RED2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
RED2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.978357 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JI20, Q3UTP1, Q8CC51, Q9JIE5 | Gene names | Adarb2, Adar3, Red2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
DSRAD_HUMAN
|
||||||
| θ value | 1.00825e-67 (rank : 5) | NC score | 0.842149 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
|
DSRAD_MOUSE
|
||||||
| θ value | 1.04338e-64 (rank : 6) | NC score | 0.835251 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
ILF3_MOUSE
|
||||||
| θ value | 1.52067e-31 (rank : 7) | NC score | 0.679098 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
ILF3_HUMAN
|
||||||
| θ value | 1.28732e-30 (rank : 8) | NC score | 0.681084 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
STAU2_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 9) | NC score | 0.448377 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CJ67, Q8BSY8, Q8CJ66, Q8R175, Q91Z19, Q9D5N7 | Gene names | Stau2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
STAU2_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 10) | NC score | 0.430855 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NUL3, Q6AHY7, Q96HM0, Q96HM1, Q9NVI5, Q9UGG6 | Gene names | STAU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
TRBP2_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 11) | NC score | 0.331498 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97473 | Gene names | Tarbp2, Prbp | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Protamine-1 RNA-binding protein) (PRM-1 RNA-binding protein). | |||||
|
TRBP2_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 12) | NC score | 0.317718 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15633, Q12878 | Gene names | TARBP2, TRBP | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Trans-activation-responsive RNA-binding protein). | |||||
|
STAU1_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 13) | NC score | 0.318058 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
STAU1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 14) | NC score | 0.315635 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z108 | Gene names | Stau1, Stau | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
PRKRA_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 15) | NC score | 0.286110 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75569, Q53G24, Q6X7T5, Q8NDK4 | Gene names | PRKRA, PACT, RAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X). | |||||
|
PRKRA_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.286701 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WTX2, Q9CZB7 | Gene names | Prkra, Rax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X) (RAX). | |||||
|
APBA1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.020868 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
TSH1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.021268 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZSZ6, O60534, Q4LE29, Q53EU4 | Gene names | TSHZ1, SDCCAG33, TSH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3) (Antigen NY-CO-33). | |||||
|
E2AK2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.029097 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19525 | Gene names | EIF2AK2, PKR, PRKR | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.008275 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.003963 (rank : 36) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
DUS1L_MOUSE
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.017637 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C2P3, Q8VCN7, Q9D124 | Gene names | Dus1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 1-like (EC 1.-.-.-). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.018113 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
BCL7C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.023875 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
GCR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.006775 (rank : 33) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04150, P04151, Q53EP5, Q6N0A4 | Gene names | NR3C1, GRL | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor (GR). | |||||
|
RAIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.016140 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5U651, Q6U676 | Gene names | RASIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-interacting protein 1 (Rain). | |||||
|
TAF5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.004860 (rank : 35) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15542, Q86UZ7, Q9Y4K5 | Gene names | TAF5, TAF2D | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 5 (Transcription initiation factor TFIID 100 kDa subunit) (TAF(II)100) (TAFII-100) (TAFII100). | |||||
|
TAF5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.004885 (rank : 34) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C092, Q8BTR2 | Gene names | Taf5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 5 (Transcription initiation factor TFIID 100 kDa subunit) (TAF(II)100) (TAFII-100) (TAFII100). | |||||
|
FLNA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.011802 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
FLNA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.011817 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
RAIN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.015392 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3U0S6, Q6GQW4 | Gene names | Rasip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-interacting protein 1 (Rain). | |||||
|
IP3KB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.007947 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27987, Q5VWL9, Q5VWM0, Q96BZ2, Q96JS1, Q9UH47 | Gene names | ITPKB | |||
|
Domain Architecture |

|
|||||
| Description | Inositol-trisphosphate 3-kinase B (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase B) (IP3K B) (IP3 3-kinase B) (IP3K-B). | |||||
|
ILF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.099025 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12905, Q5SR10, Q5SR11, Q7L7R3, Q9BWD4, Q9P1N0 | Gene names | ILF2, NF45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin enhancer-binding factor 2 (Nuclear factor of activated T- cells 45 kDa). | |||||
|
ILF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.099025 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXY6, Q3U083, Q5RKG0, Q8CCY9, Q99KS3 | Gene names | Ilf2, Nf45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin enhancer-binding factor 2 (Nuclear factor of activated T- cells 45 kDa). | |||||
|
SEPN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.055427 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9NZV5, Q6PI70, Q969F6, Q9NUI6 | Gene names | SEPN1, SELN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein N precursor. | |||||
|
ZBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.054357 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY24, Q8VE02, Q9D8B9 | Gene names | Zbp1, Dlm1 | |||
|
Domain Architecture |
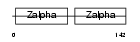
|
|||||
| Description | Z-DNA-binding protein 1 (Tumor stroma and activated macrophage protein DLM-1). | |||||
|
RED1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91ZS8, Q8K3X1, Q91ZS6, Q91ZS7, Q91ZS9, Q99MU8 | Gene names | Adarb1, Adar2, Red1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
RED1_HUMAN
|
||||||
| NC score | 0.994252 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P78563, O00395, O00465, O00691, O00692, P78555, Q8NFD1 | Gene names | ADARB1, ADAR2, DRADA2, RED1 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
RED2_HUMAN
|
||||||
| NC score | 0.982121 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NS39 | Gene names | ADARB2, ADAR3, RED2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
RED2_MOUSE
|
||||||
| NC score | 0.978357 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JI20, Q3UTP1, Q8CC51, Q9JIE5 | Gene names | Adarb2, Adar3, Red2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
DSRAD_HUMAN
|
||||||
| NC score | 0.842149 (rank : 5) | θ value | 1.00825e-67 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
|
DSRAD_MOUSE
|
||||||
| NC score | 0.835251 (rank : 6) | θ value | 1.04338e-64 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
ILF3_HUMAN
|
||||||
| NC score | 0.681084 (rank : 7) | θ value | 1.28732e-30 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
ILF3_MOUSE
|
||||||
| NC score | 0.679098 (rank : 8) | θ value | 1.52067e-31 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
STAU2_MOUSE
|
||||||
| NC score | 0.448377 (rank : 9) | θ value | 7.34386e-10 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CJ67, Q8BSY8, Q8CJ66, Q8R175, Q91Z19, Q9D5N7 | Gene names | Stau2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
STAU2_HUMAN
|
||||||
| NC score | 0.430855 (rank : 10) | θ value | 2.79066e-09 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NUL3, Q6AHY7, Q96HM0, Q96HM1, Q9NVI5, Q9UGG6 | Gene names | STAU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
TRBP2_MOUSE
|
||||||
| NC score | 0.331498 (rank : 11) | θ value | 8.40245e-06 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97473 | Gene names | Tarbp2, Prbp | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Protamine-1 RNA-binding protein) (PRM-1 RNA-binding protein). | |||||
|
STAU1_HUMAN
|
||||||
| NC score | 0.318058 (rank : 12) | θ value | 0.000270298 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
TRBP2_HUMAN
|
||||||
| NC score | 0.317718 (rank : 13) | θ value | 1.09739e-05 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15633, Q12878 | Gene names | TARBP2, TRBP | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Trans-activation-responsive RNA-binding protein). | |||||
|
STAU1_MOUSE
|
||||||
| NC score | 0.315635 (rank : 14) | θ value | 0.00134147 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z108 | Gene names | Stau1, Stau | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
PRKRA_MOUSE
|
||||||
| NC score | 0.286701 (rank : 15) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WTX2, Q9CZB7 | Gene names | Prkra, Rax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X) (RAX). | |||||
|
PRKRA_HUMAN
|
||||||
| NC score | 0.286110 (rank : 16) | θ value | 0.0961366 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75569, Q53G24, Q6X7T5, Q8NDK4 | Gene names | PRKRA, PACT, RAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X). | |||||
|
ILF2_HUMAN
|
||||||
| NC score | 0.099025 (rank : 17) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12905, Q5SR10, Q5SR11, Q7L7R3, Q9BWD4, Q9P1N0 | Gene names | ILF2, NF45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin enhancer-binding factor 2 (Nuclear factor of activated T- cells 45 kDa). | |||||
|
ILF2_MOUSE
|
||||||
| NC score | 0.099025 (rank : 18) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXY6, Q3U083, Q5RKG0, Q8CCY9, Q99KS3 | Gene names | Ilf2, Nf45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin enhancer-binding factor 2 (Nuclear factor of activated T- cells 45 kDa). | |||||
|
SEPN1_HUMAN
|
||||||
| NC score | 0.055427 (rank : 19) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9NZV5, Q6PI70, Q969F6, Q9NUI6 | Gene names | SEPN1, SELN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein N precursor. | |||||
|
ZBP1_MOUSE
|
||||||
| NC score | 0.054357 (rank : 20) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY24, Q8VE02, Q9D8B9 | Gene names | Zbp1, Dlm1 | |||
|
Domain Architecture |

|
|||||
| Description | Z-DNA-binding protein 1 (Tumor stroma and activated macrophage protein DLM-1). | |||||
|
E2AK2_HUMAN
|
||||||
| NC score | 0.029097 (rank : 21) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19525 | Gene names | EIF2AK2, PKR, PRKR | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase). | |||||
|
BCL7C_MOUSE
|
||||||
| NC score | 0.023875 (rank : 22) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
TSH1_HUMAN
|
||||||
| NC score | 0.021268 (rank : 23) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZSZ6, O60534, Q4LE29, Q53EU4 | Gene names | TSHZ1, SDCCAG33, TSH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3) (Antigen NY-CO-33). | |||||
|
APBA1_HUMAN
|
||||||
| NC score | 0.020868 (rank : 24) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.018113 (rank : 25) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
DUS1L_MOUSE
|
||||||
| NC score | 0.017637 (rank : 26) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C2P3, Q8VCN7, Q9D124 | Gene names | Dus1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 1-like (EC 1.-.-.-). | |||||
|
RAIN_HUMAN
|
||||||
| NC score | 0.016140 (rank : 27) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5U651, Q6U676 | Gene names | RASIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-interacting protein 1 (Rain). | |||||
|
RAIN_MOUSE
|
||||||
| NC score | 0.015392 (rank : 28) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3U0S6, Q6GQW4 | Gene names | Rasip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-interacting protein 1 (Rain). | |||||
|
FLNA_MOUSE
|
||||||
| NC score | 0.011817 (rank : 29) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
FLNA_HUMAN
|
||||||
| NC score | 0.011802 (rank : 30) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.008275 (rank : 31) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
IP3KB_HUMAN
|
||||||
| NC score | 0.007947 (rank : 32) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27987, Q5VWL9, Q5VWM0, Q96BZ2, Q96JS1, Q9UH47 | Gene names | ITPKB | |||
|
Domain Architecture |

|
|||||
| Description | Inositol-trisphosphate 3-kinase B (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase B) (IP3K B) (IP3 3-kinase B) (IP3K-B). | |||||
|
GCR_HUMAN
|
||||||
| NC score | 0.006775 (rank : 33) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04150, P04151, Q53EP5, Q6N0A4 | Gene names | NR3C1, GRL | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor (GR). | |||||
|
TAF5_MOUSE
|
||||||
| NC score | 0.004885 (rank : 34) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C092, Q8BTR2 | Gene names | Taf5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 5 (Transcription initiation factor TFIID 100 kDa subunit) (TAF(II)100) (TAFII-100) (TAFII100). | |||||
|
TAF5_HUMAN
|
||||||
| NC score | 0.004860 (rank : 35) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15542, Q86UZ7, Q9Y4K5 | Gene names | TAF5, TAF2D | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 5 (Transcription initiation factor TFIID 100 kDa subunit) (TAF(II)100) (TAFII-100) (TAFII100). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.003963 (rank : 36) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||