Please be patient as the page loads
|
STAU1_HUMAN
|
||||||
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STAU1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
STAU1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993254 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z108 | Gene names | Stau1, Stau | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
STAU2_MOUSE
|
||||||
| θ value | 1.18275e-92 (rank : 3) | NC score | 0.929688 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CJ67, Q8BSY8, Q8CJ66, Q8R175, Q91Z19, Q9D5N7 | Gene names | Stau2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
STAU2_HUMAN
|
||||||
| θ value | 7.66637e-92 (rank : 4) | NC score | 0.930971 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NUL3, Q6AHY7, Q96HM0, Q96HM1, Q9NVI5, Q9UGG6 | Gene names | STAU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
PRKRA_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 5) | NC score | 0.583810 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WTX2, Q9CZB7 | Gene names | Prkra, Rax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X) (RAX). | |||||
|
PRKRA_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 6) | NC score | 0.581198 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75569, Q53G24, Q6X7T5, Q8NDK4 | Gene names | PRKRA, PACT, RAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X). | |||||
|
TRBP2_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 7) | NC score | 0.508696 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15633, Q12878 | Gene names | TARBP2, TRBP | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Trans-activation-responsive RNA-binding protein). | |||||
|
TRBP2_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 8) | NC score | 0.491916 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97473 | Gene names | Tarbp2, Prbp | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Protamine-1 RNA-binding protein) (PRM-1 RNA-binding protein). | |||||
|
ILF3_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 9) | NC score | 0.344985 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
RED1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 10) | NC score | 0.318899 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78563, O00395, O00465, O00691, O00692, P78555, Q8NFD1 | Gene names | ADARB1, ADAR2, DRADA2, RED1 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
RED1_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 11) | NC score | 0.318058 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91ZS8, Q8K3X1, Q91ZS6, Q91ZS7, Q91ZS9, Q99MU8 | Gene names | Adarb1, Adar2, Red1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
ILF3_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 12) | NC score | 0.334482 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
DSRAD_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 13) | NC score | 0.276470 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
|
RED2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 14) | NC score | 0.266698 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NS39 | Gene names | ADARB2, ADAR3, RED2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 15) | NC score | 0.061239 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
RED2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.268206 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JI20, Q3UTP1, Q8CC51, Q9JIE5 | Gene names | Adarb2, Adar3, Red2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
BMP2K_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.020894 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NSY1, O94791, Q8IYF2, Q8N2G7, Q8NHG9, Q9NTG8 | Gene names | BMP2K, BIKE | |||
|
Domain Architecture |
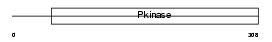
|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
E2AK2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.024482 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q03963, Q61742, Q62026 | Gene names | Eif2ak2, Pkr, Prkr, Tik | |||
|
Domain Architecture |
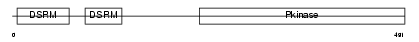
|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase) (Serine/threonine-protein kinase TIK). | |||||
|
E2AK2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.031818 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19525 | Gene names | EIF2AK2, PKR, PRKR | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.019262 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
DHX9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.024782 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
ZN500_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.001327 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60304, Q96CQ8, Q9BTG0 | Gene names | ZNF500, KIAA0557 | |||
|
Domain Architecture |
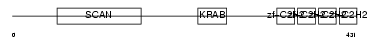
|
|||||
| Description | Zinc finger protein 500. | |||||
|
DCP1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.051853 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YD3, Q6NZE3 | Gene names | Dcp1a, Mitc1, Smif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (MAD homolog 4-interacting transcription coactivator 1) (Smad4-interacting transcriptional co-activator). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.053745 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
SMAP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.022568 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
DSRAD_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.251661 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
SRPR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.024094 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBG7, Q8VC45, Q921H1 | Gene names | Srpr | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle receptor subunit alpha (SR-alpha) (Docking protein alpha) (DP-alpha). | |||||
|
DHX9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.019058 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
ANK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.007507 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ANK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.007048 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
CEBPB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.019105 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
DCP1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.052416 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
FYV1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.009053 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
IPO4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.011979 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VI75, Q3TBW0, Q99J52 | Gene names | Ipo4, Imp4a, Ranbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-4 (Importin 4a) (Imp4a) (Ran-binding protein 4) (RanBP4). | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.026584 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
ZN432_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | -0.000851 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94892 | Gene names | ZNF432, KIAA0798 | |||
|
Domain Architecture |
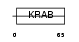
|
|||||
| Description | Zinc finger protein 432. | |||||
|
CEBPB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.016056 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28033 | Gene names | Cebpb | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Interleukin-6- dependent-binding protein) (IL-6DBP) (Liver-enriched transcriptional activator) (LAP) (AGP/EBP). | |||||
|
DEN1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.007487 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
DGCR8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.041940 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WYQ5, Q6DCB2, Q6MZE9, Q6Y2L0, Q96G39, Q96GP8, Q9H6L8, Q9H6T7, Q9NRW2 | Gene names | DGCR8, C22orf12, DGCRK6 | |||
|
Domain Architecture |

|
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8). | |||||
|
DGCR8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.041936 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQM6 | Gene names | Dgcr8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8 homolog) (Gy1). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.004775 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
MARK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.001361 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.005842 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.015943 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TOP2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.007621 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01320 | Gene names | Top2a, Top-2, Top2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
STAU1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
STAU1_MOUSE
|
||||||
| NC score | 0.993254 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z108 | Gene names | Stau1, Stau | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
STAU2_HUMAN
|
||||||
| NC score | 0.930971 (rank : 3) | θ value | 7.66637e-92 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NUL3, Q6AHY7, Q96HM0, Q96HM1, Q9NVI5, Q9UGG6 | Gene names | STAU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
STAU2_MOUSE
|
||||||
| NC score | 0.929688 (rank : 4) | θ value | 1.18275e-92 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CJ67, Q8BSY8, Q8CJ66, Q8R175, Q91Z19, Q9D5N7 | Gene names | Stau2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
PRKRA_MOUSE
|
||||||
| NC score | 0.583810 (rank : 5) | θ value | 3.18553e-13 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WTX2, Q9CZB7 | Gene names | Prkra, Rax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X) (RAX). | |||||
|
PRKRA_HUMAN
|
||||||
| NC score | 0.581198 (rank : 6) | θ value | 4.16044e-13 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75569, Q53G24, Q6X7T5, Q8NDK4 | Gene names | PRKRA, PACT, RAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X). | |||||
|
TRBP2_HUMAN
|
||||||
| NC score | 0.508696 (rank : 7) | θ value | 4.76016e-09 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15633, Q12878 | Gene names | TARBP2, TRBP | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Trans-activation-responsive RNA-binding protein). | |||||
|
TRBP2_MOUSE
|
||||||
| NC score | 0.491916 (rank : 8) | θ value | 1.17247e-07 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97473 | Gene names | Tarbp2, Prbp | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Protamine-1 RNA-binding protein) (PRM-1 RNA-binding protein). | |||||
|
ILF3_MOUSE
|
||||||
| NC score | 0.344985 (rank : 9) | θ value | 0.00020696 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
ILF3_HUMAN
|
||||||
| NC score | 0.334482 (rank : 10) | θ value | 0.00035302 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
RED1_HUMAN
|
||||||
| NC score | 0.318899 (rank : 11) | θ value | 0.00020696 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78563, O00395, O00465, O00691, O00692, P78555, Q8NFD1 | Gene names | ADARB1, ADAR2, DRADA2, RED1 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
RED1_MOUSE
|
||||||
| NC score | 0.318058 (rank : 12) | θ value | 0.000270298 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91ZS8, Q8K3X1, Q91ZS6, Q91ZS7, Q91ZS9, Q99MU8 | Gene names | Adarb1, Adar2, Red1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
DSRAD_HUMAN
|
||||||
| NC score | 0.276470 (rank : 13) | θ value | 0.00869519 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
|
RED2_MOUSE
|
||||||
| NC score | 0.268206 (rank : 14) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JI20, Q3UTP1, Q8CC51, Q9JIE5 | Gene names | Adarb2, Adar3, Red2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
RED2_HUMAN
|
||||||
| NC score | 0.266698 (rank : 15) | θ value | 0.00869519 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NS39 | Gene names | ADARB2, ADAR3, RED2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
DSRAD_MOUSE
|
||||||
| NC score | 0.251661 (rank : 16) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.061239 (rank : 17) | θ value | 0.0193708 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.053745 (rank : 18) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
DCP1A_HUMAN
|
||||||
| NC score | 0.052416 (rank : 19) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
DCP1A_MOUSE
|
||||||
| NC score | 0.051853 (rank : 20) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YD3, Q6NZE3 | Gene names | Dcp1a, Mitc1, Smif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (MAD homolog 4-interacting transcription coactivator 1) (Smad4-interacting transcriptional co-activator). | |||||
|
DGCR8_HUMAN
|
||||||
| NC score | 0.041940 (rank : 21) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WYQ5, Q6DCB2, Q6MZE9, Q6Y2L0, Q96G39, Q96GP8, Q9H6L8, Q9H6T7, Q9NRW2 | Gene names | DGCR8, C22orf12, DGCRK6 | |||
|
Domain Architecture |

|
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8). | |||||
|
DGCR8_MOUSE
|
||||||
| NC score | 0.041936 (rank : 22) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQM6 | Gene names | Dgcr8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8 homolog) (Gy1). | |||||
|
E2AK2_HUMAN
|
||||||
| NC score | 0.031818 (rank : 23) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19525 | Gene names | EIF2AK2, PKR, PRKR | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase). | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.026584 (rank : 24) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
DHX9_HUMAN
|
||||||
| NC score | 0.024782 (rank : 25) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
E2AK2_MOUSE
|
||||||
| NC score | 0.024482 (rank : 26) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q03963, Q61742, Q62026 | Gene names | Eif2ak2, Pkr, Prkr, Tik | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase) (Serine/threonine-protein kinase TIK). | |||||
|
SRPR_MOUSE
|
||||||
| NC score | 0.024094 (rank : 27) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBG7, Q8VC45, Q921H1 | Gene names | Srpr | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle receptor subunit alpha (SR-alpha) (Docking protein alpha) (DP-alpha). | |||||
|
SMAP1_MOUSE
|
||||||
| NC score | 0.022568 (rank : 28) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
BMP2K_HUMAN
|
||||||
| NC score | 0.020894 (rank : 29) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NSY1, O94791, Q8IYF2, Q8N2G7, Q8NHG9, Q9NTG8 | Gene names | BMP2K, BIKE | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.019262 (rank : 30) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
CEBPB_HUMAN
|
||||||
| NC score | 0.019105 (rank : 31) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
DHX9_MOUSE
|
||||||
| NC score | 0.019058 (rank : 32) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
CEBPB_MOUSE
|
||||||
| NC score | 0.016056 (rank : 33) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28033 | Gene names | Cebpb | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Interleukin-6- dependent-binding protein) (IL-6DBP) (Liver-enriched transcriptional activator) (LAP) (AGP/EBP). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.015943 (rank : 34) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
IPO4_MOUSE
|
||||||
| NC score | 0.011979 (rank : 35) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VI75, Q3TBW0, Q99J52 | Gene names | Ipo4, Imp4a, Ranbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-4 (Importin 4a) (Imp4a) (Ran-binding protein 4) (RanBP4). | |||||
|
FYV1_HUMAN
|
||||||
| NC score | 0.009053 (rank : 36) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
TOP2A_MOUSE
|
||||||
| NC score | 0.007621 (rank : 37) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01320 | Gene names | Top2a, Top-2, Top2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
ANK1_HUMAN
|
||||||
| NC score | 0.007507 (rank : 38) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
DEN1A_HUMAN
|
||||||
| NC score | 0.007487 (rank : 39) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
ANK1_MOUSE
|
||||||
| NC score | 0.007048 (rank : 40) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.005842 (rank : 41) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.004775 (rank : 42) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
MARK2_HUMAN
|
||||||
| NC score | 0.001361 (rank : 43) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
ZN500_HUMAN
|
||||||
| NC score | 0.001327 (rank : 44) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60304, Q96CQ8, Q9BTG0 | Gene names | ZNF500, KIAA0557 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 500. | |||||
|
ZN432_HUMAN
|
||||||
| NC score | -0.000851 (rank : 45) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94892 | Gene names | ZNF432, KIAA0798 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 432. | |||||