Please be patient as the page loads
|
CABL1_HUMAN
|
||||||
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CABL1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
CABL1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.984846 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ESJ1, Q9EPR8 | Gene names | Cables1, Cables | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
CABL2_HUMAN
|
||||||
| θ value | 1.21265e-121 (rank : 3) | NC score | 0.944970 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
CABL2_MOUSE
|
||||||
| θ value | 1.81207e-117 (rank : 4) | NC score | 0.947916 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K3M5 | Gene names | Cables2 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 5) | NC score | 0.065270 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
SUSD4_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 6) | NC score | 0.050289 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
SDC1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 7) | NC score | 0.056193 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18828, Q62278, Q9WVD2 | Gene names | Sdc1, Synd-1, Synd1 | |||
|
Domain Architecture |
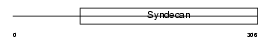
|
|||||
| Description | Syndecan-1 precursor (SYND1). | |||||
|
NOS3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 8) | NC score | 0.034813 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70313, O55056 | Gene names | Nos3, Ecnos | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, endothelial (EC 1.14.13.39) (EC-NOS) (NOS type III) (NOSIII) (Endothelial NOS) (eNOS) (Constitutive NOS) (cNOS). | |||||
|
DGCR8_HUMAN
|
||||||
| θ value | 0.62314 (rank : 9) | NC score | 0.058096 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WYQ5, Q6DCB2, Q6MZE9, Q6Y2L0, Q96G39, Q96GP8, Q9H6L8, Q9H6T7, Q9NRW2 | Gene names | DGCR8, C22orf12, DGCRK6 | |||
|
Domain Architecture |

|
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.027407 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
PAF49_MOUSE
|
||||||
| θ value | 0.62314 (rank : 11) | NC score | 0.045380 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76KJ5, Q8K0Y9 | Gene names | Cd3eap, Ase1, Paf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein). | |||||
|
SUSD4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.045168 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VX71, Q6UX62, Q9BSR0, Q9NWG0 | Gene names | SUSD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.028147 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
TMUB1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.039378 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JMG3 | Gene names | Tmub1 | |||
|
Domain Architecture |
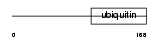
|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.043623 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.043556 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.033470 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
CCNT2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.061522 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.023492 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
NOS3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.030756 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29474, Q13662, Q14251, Q14434 | Gene names | NOS3 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, endothelial (EC 1.14.13.39) (EC-NOS) (NOS type III) (NOSIII) (Endothelial NOS) (eNOS) (Constitutive NOS) (cNOS). | |||||
|
DGCR8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.047129 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQM6 | Gene names | Dgcr8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8 homolog) (Gy1). | |||||
|
SMOO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.019725 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.010856 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
SCNND_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.016820 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51172, Q5T7L3, Q8NA24 | Gene names | SCNN1D, DNACH | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit delta (Epithelial Na+ channel subunit delta) (Delta ENaC) (Nonvoltage-gated sodium channel 1 subunit delta) (SCNED) (Delta NaCH). | |||||
|
PKHG4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.017491 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
ZDHC8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.012202 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULC8, Q6ICL1, Q6ZNF5, Q7Z6L9 | Gene names | ZDHHC8, KIAA1292, ZDHHCL1, ZNF378 | |||
|
Domain Architecture |
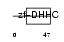
|
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8) (Zinc finger protein 378). | |||||
|
GASP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.013484 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
SAKS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.018008 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q922Y1, Q3UCP8 | Gene names | Saks1, D19Ertd721e | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein) (mY33K) (Protein 2B28). | |||||
|
CCNC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.058419 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P24863, Q9H543 | Gene names | CCNC | |||
|
Domain Architecture |
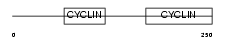
|
|||||
| Description | Cyclin-C. | |||||
|
CCNC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.056377 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62447, Q9WUZ4 | Gene names | Ccnc | |||
|
Domain Architecture |
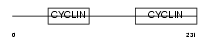
|
|||||
| Description | Cyclin-C. | |||||
|
CPSM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.022483 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31327, O43774, Q7Z5I5 | Gene names | CPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
IBP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.012117 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
SPHK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.015348 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JIA7, Q91VA9, Q9DBH6 | Gene names | Sphk2 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
ZHX2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.010358 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.011560 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.021597 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CPSM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.016789 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
CABL1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
CABL1_MOUSE
|
||||||
| NC score | 0.984846 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ESJ1, Q9EPR8 | Gene names | Cables1, Cables | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
CABL2_MOUSE
|
||||||
| NC score | 0.947916 (rank : 3) | θ value | 1.81207e-117 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K3M5 | Gene names | Cables2 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
CABL2_HUMAN
|
||||||
| NC score | 0.944970 (rank : 4) | θ value | 1.21265e-121 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.065270 (rank : 5) | θ value | 0.000461057 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
CCNT2_HUMAN
|
||||||
| NC score | 0.061522 (rank : 6) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
CCNC_HUMAN
|
||||||
| NC score | 0.058419 (rank : 7) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P24863, Q9H543 | Gene names | CCNC | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-C. | |||||
|
DGCR8_HUMAN
|
||||||
| NC score | 0.058096 (rank : 8) | θ value | 0.62314 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WYQ5, Q6DCB2, Q6MZE9, Q6Y2L0, Q96G39, Q96GP8, Q9H6L8, Q9H6T7, Q9NRW2 | Gene names | DGCR8, C22orf12, DGCRK6 | |||
|
Domain Architecture |

|
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8). | |||||
|
CCNC_MOUSE
|
||||||
| NC score | 0.056377 (rank : 9) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62447, Q9WUZ4 | Gene names | Ccnc | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-C. | |||||
|
SDC1_MOUSE
|
||||||
| NC score | 0.056193 (rank : 10) | θ value | 0.163984 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18828, Q62278, Q9WVD2 | Gene names | Sdc1, Synd-1, Synd1 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-1 precursor (SYND1). | |||||
|
SUSD4_MOUSE
|
||||||
| NC score | 0.050289 (rank : 11) | θ value | 0.0961366 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
DGCR8_MOUSE
|
||||||
| NC score | 0.047129 (rank : 12) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQM6 | Gene names | Dgcr8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DGCR8 protein (DiGeorge syndrome critical region 8 homolog) (Gy1). | |||||
|
PAF49_MOUSE
|
||||||
| NC score | 0.045380 (rank : 13) | θ value | 0.62314 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76KJ5, Q8K0Y9 | Gene names | Cd3eap, Ase1, Paf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein). | |||||
|
SUSD4_HUMAN
|
||||||
| NC score | 0.045168 (rank : 14) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VX71, Q6UX62, Q9BSR0, Q9NWG0 | Gene names | SUSD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.043623 (rank : 15) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.043556 (rank : 16) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
TMUB1_MOUSE
|
||||||
| NC score | 0.039378 (rank : 17) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JMG3 | Gene names | Tmub1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1. | |||||
|
NOS3_MOUSE
|
||||||
| NC score | 0.034813 (rank : 18) | θ value | 0.365318 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70313, O55056 | Gene names | Nos3, Ecnos | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, endothelial (EC 1.14.13.39) (EC-NOS) (NOS type III) (NOSIII) (Endothelial NOS) (eNOS) (Constitutive NOS) (cNOS). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.033470 (rank : 19) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
NOS3_HUMAN
|
||||||
| NC score | 0.030756 (rank : 20) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29474, Q13662, Q14251, Q14434 | Gene names | NOS3 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, endothelial (EC 1.14.13.39) (EC-NOS) (NOS type III) (NOSIII) (Endothelial NOS) (eNOS) (Constitutive NOS) (cNOS). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.028147 (rank : 21) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.027407 (rank : 22) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.023492 (rank : 23) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
CPSM_HUMAN
|
||||||
| NC score | 0.022483 (rank : 24) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31327, O43774, Q7Z5I5 | Gene names | CPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.021597 (rank : 25) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.019725 (rank : 26) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
SAKS1_MOUSE
|
||||||
| NC score | 0.018008 (rank : 27) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q922Y1, Q3UCP8 | Gene names | Saks1, D19Ertd721e | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein) (mY33K) (Protein 2B28). | |||||
|
PKHG4_HUMAN
|
||||||
| NC score | 0.017491 (rank : 28) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
SCNND_HUMAN
|
||||||
| NC score | 0.016820 (rank : 29) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51172, Q5T7L3, Q8NA24 | Gene names | SCNN1D, DNACH | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit delta (Epithelial Na+ channel subunit delta) (Delta ENaC) (Nonvoltage-gated sodium channel 1 subunit delta) (SCNED) (Delta NaCH). | |||||
|
CPSM_MOUSE
|
||||||
| NC score | 0.016789 (rank : 30) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
SPHK2_MOUSE
|
||||||
| NC score | 0.015348 (rank : 31) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JIA7, Q91VA9, Q9DBH6 | Gene names | Sphk2 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
GASP2_MOUSE
|
||||||
| NC score | 0.013484 (rank : 32) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
ZDHC8_HUMAN
|
||||||
| NC score | 0.012202 (rank : 33) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULC8, Q6ICL1, Q6ZNF5, Q7Z6L9 | Gene names | ZDHHC8, KIAA1292, ZDHHCL1, ZNF378 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8) (Zinc finger protein 378). | |||||
|
IBP1_MOUSE
|
||||||
| NC score | 0.012117 (rank : 34) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.011560 (rank : 35) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.010856 (rank : 36) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
ZHX2_MOUSE
|
||||||
| NC score | 0.010358 (rank : 37) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||