Please be patient as the page loads
|
CCNT2_HUMAN
|
||||||
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CCNT2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
CCNT1_MOUSE
|
||||||
| θ value | 8.35901e-139 (rank : 2) | NC score | 0.958644 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT1_HUMAN
|
||||||
| θ value | 1.02182e-136 (rank : 3) | NC score | 0.957187 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60563, O60581 | Gene names | CCNT1 | |||
|
Domain Architecture |
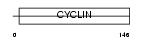
|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNK_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 4) | NC score | 0.821365 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 4.42448e-31 (rank : 5) | NC score | 0.709457 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CCNL1_MOUSE
|
||||||
| θ value | 1.29031e-22 (rank : 6) | NC score | 0.723499 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
CCNL2_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 7) | NC score | 0.724367 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
CCNL2_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 8) | NC score | 0.716803 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
CCNL1_HUMAN
|
||||||
| θ value | 3.17815e-21 (rank : 9) | NC score | 0.720383 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CCNC_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 10) | NC score | 0.656539 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P24863, Q9H543 | Gene names | CCNC | |||
|
Domain Architecture |
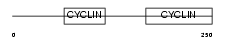
|
|||||
| Description | Cyclin-C. | |||||
|
CCNC_MOUSE
|
||||||
| θ value | 7.84624e-12 (rank : 11) | NC score | 0.636911 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62447, Q9WUZ4 | Gene names | Ccnc | |||
|
Domain Architecture |
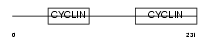
|
|||||
| Description | Cyclin-C. | |||||
|
CCNH_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 12) | NC score | 0.338186 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51946, Q8TBL9 | Gene names | CCNH | |||
|
Domain Architecture |
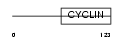
|
|||||
| Description | Cyclin-H (MO15-associated protein) (p37) (p34). | |||||
|
CCNH_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 13) | NC score | 0.306682 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61458, Q9CVJ0, Q9JHV7 | Gene names | Ccnh | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-H. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 14) | NC score | 0.087146 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
MANS1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.056664 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H8J5, Q8NEC1 | Gene names | MANSC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MANSC domain-containing protein 1 precursor. | |||||
|
CCNA2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.093055 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
JARD2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.035166 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
WDR20_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.029413 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TBZ3, Q8WXX2, Q9UF86 | Gene names | WDR20 | |||
|
Domain Architecture |
|
|||||
| Description | WD repeat protein 20 (DMR protein). | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.062180 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CDKL5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.005011 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.040299 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
CABL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.068007 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESJ1, Q9EPR8 | Gene names | Cables1, Cables | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
CABL2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.066346 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K3M5 | Gene names | Cables2 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
SMBP2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.028419 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
PAPPA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.040676 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R4K8, Q80VW3, Q80YY1, Q8K423, Q8R4K7, Q9ES06, Q9JK57 | Gene names | Pappa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.017646 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
X3CL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.032166 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35188, O35933 | Gene names | Cx3cl1, Cx3c, Fkn, Scyd1 | |||
|
Domain Architecture |
|
|||||
| Description | Fractalkine precursor (CX3CL1) (Neurotactin) (CX3C membrane-anchored chemokine) (Small inducible cytokine D1). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.026996 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
CABL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.061522 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
DPOLZ_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.036845 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.038485 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.032038 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
ATS12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.007320 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58397, Q6UWL3 | Gene names | ADAMTS12 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
CN093_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.028497 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H972, Q86SE6, Q96CF7, Q9HA68 | Gene names | C14orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf93 precursor. | |||||
|
DPOLZ_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.033761 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.025661 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.023069 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
PAPPA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.034754 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.043639 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
BCLF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.027048 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
CABL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.062254 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
EVI1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.000582 (rank : 86) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
MEPE_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.042051 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQ76 | Gene names | MEPE | |||
|
Domain Architecture |
|
|||||
| Description | Matrix extracellular phosphoglycoprotein precursor (Osteoblast/osteocyte factor 45) (OF45). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.021343 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
RBP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.016020 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
E41L3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.007886 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
MTR1L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.001455 (rank : 85) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.009836 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
SEMG2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.031971 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
GRP75_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.011148 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
GRP75_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.011183 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
LIRA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.007026 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6C8, O15469, O15470, O75016, Q8N151, Q8N154, Q8NHJ1, Q8NHJ2, Q8NHJ3, Q8NHJ4 | Gene names | LILRA3, ILT6, LIR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 3 precursor (Leukocyte immunoglobulin-like receptor 4) (LIR-4) (Immunoglobulin- like transcript 6) (ILT-6) (Monocyte inhibitory receptor HM43/HM31) (CD85e antigen). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.009236 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.017651 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.021983 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.035557 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.032475 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
ARD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.006572 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 621 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36406, Q9BZY4, Q9BZY5 | Gene names | TRIM23, ARD1, ARFD1, RNF46 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein ARD-1 (ADP-ribosylation factor domain protein 1) (Tripartite motif-containing protein 23) (RING finger protein 46). | |||||
|
ARD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.006816 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGX0, Q8C2B6, Q8CDA4, Q8CDA7 | Gene names | Trim23, Arfd1 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein ARD-1 (ADP-ribosylation factor domain protein 1) (Tripartite motif-containing protein 23). | |||||
|
CENPT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.019049 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96BT3, Q96I29, Q96IC6, Q96NK9, Q9H901 | Gene names | CENPT, C16orf56, ICEN22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T) (Interphase centromere complex protein 22). | |||||
|
COHA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.017071 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
HORN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.033155 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.028884 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LUZP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.008538 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
PLK4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | -0.000678 (rank : 87) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00444, Q8IYF0, Q96Q95 | Gene names | PLK4, SAK, STK18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PLK4 (EC 2.7.11.21) (Polo-like kinase 4) (PLK-4) (Serine/threonine-protein kinase Sak) (Serine/threonine- protein kinase 18). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.004364 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
ZN292_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.009597 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
ARHGC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.006245 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.090341 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.005141 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.013148 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
PROM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.014210 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43490 | Gene names | PROM1, PROML1 | |||
|
Domain Architecture |

|
|||||
| Description | Prominin-1 precursor (Prominin-like protein 1) (Antigen AC133) (CD133 antigen). | |||||
|
SIN3A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.009519 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.026085 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TEX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.014493 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZPJ0 | Gene names | Tex2, Kiaa1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
ZCHC7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.019474 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N3Z6, Q5T0Q8, Q5T0Q9, Q5T0R0, Q8N2M1, Q8N4J2, Q8TBK8, Q9H648, Q9P0F0 | Gene names | ZCCHC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 7. | |||||
|
JADE3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.007494 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
K1196_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.005850 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96KM6, Q9ULM4 | Gene names | KIAA1196 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1196. | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.010812 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MAML1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.009370 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.011507 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PRS23_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.011814 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95084 | Gene names | PRSS23, ZSIG13 | |||
|
Domain Architecture |
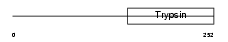
|
|||||
| Description | Serine protease 23 precursor (EC 3.4.21.-) (Putative secreted protein ZSIG13). | |||||
|
RNF6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.011107 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y252, Q9UF41 | Gene names | RNF6 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 6 (RING-H2 protein). | |||||
|
ZN148_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | -0.001122 (rank : 88) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UQR1, O00389, O43591 | Gene names | ZNF148, ZBP89 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89). | |||||
|
CCNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.067213 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.062255 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |
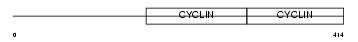
|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.056445 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.057535 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CCNT2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
CCNT1_MOUSE
|
||||||
| NC score | 0.958644 (rank : 2) | θ value | 8.35901e-139 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT1_HUMAN
|
||||||
| NC score | 0.957187 (rank : 3) | θ value | 1.02182e-136 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60563, O60581 | Gene names | CCNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNK_HUMAN
|
||||||
| NC score | 0.821365 (rank : 4) | θ value | 1.52067e-31 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNL2_MOUSE
|
||||||
| NC score | 0.724367 (rank : 5) | θ value | 3.75424e-22 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
CCNL1_MOUSE
|
||||||
| NC score | 0.723499 (rank : 6) | θ value | 1.29031e-22 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
CCNL1_HUMAN
|
||||||
| NC score | 0.720383 (rank : 7) | θ value | 3.17815e-21 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CCNL2_HUMAN
|
||||||
| NC score | 0.716803 (rank : 8) | θ value | 1.86321e-21 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.709457 (rank : 9) | θ value | 4.42448e-31 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CCNC_HUMAN
|
||||||
| NC score | 0.656539 (rank : 10) | θ value | 4.16044e-13 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P24863, Q9H543 | Gene names | CCNC | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-C. | |||||
|
CCNC_MOUSE
|
||||||
| NC score | 0.636911 (rank : 11) | θ value | 7.84624e-12 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62447, Q9WUZ4 | Gene names | Ccnc | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-C. | |||||
|
CCNH_HUMAN
|
||||||
| NC score | 0.338186 (rank : 12) | θ value | 0.000786445 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51946, Q8TBL9 | Gene names | CCNH | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-H (MO15-associated protein) (p37) (p34). | |||||
|
CCNH_MOUSE
|
||||||
| NC score | 0.306682 (rank : 13) | θ value | 0.00298849 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61458, Q9CVJ0, Q9JHV7 | Gene names | Ccnh | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-H. | |||||
|
CCNA2_HUMAN
|
||||||
| NC score | 0.093055 (rank : 14) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.090341 (rank : 15) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.087146 (rank : 16) | θ value | 0.00869519 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
CABL1_MOUSE
|
||||||
| NC score | 0.068007 (rank : 17) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESJ1, Q9EPR8 | Gene names | Cables1, Cables | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
CCNA1_HUMAN
|
||||||
| NC score | 0.067213 (rank : 18) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CABL2_MOUSE
|
||||||
| NC score | 0.066346 (rank : 19) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K3M5 | Gene names | Cables2 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
CCNA1_MOUSE
|
||||||
| NC score | 0.062255 (rank : 20) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CABL2_HUMAN
|
||||||
| NC score | 0.062254 (rank : 21) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.062180 (rank : 22) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CABL1_HUMAN
|
||||||
| NC score | 0.061522 (rank : 23) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.057535 (rank : 24) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
MANS1_HUMAN
|
||||||
| NC score | 0.056664 (rank : 25) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H8J5, Q8NEC1 | Gene names | MANSC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MANSC domain-containing protein 1 precursor. | |||||
|
CCNA2_MOUSE
|
||||||
| NC score | 0.056445 (rank : 26) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.043639 (rank : 27) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
MEPE_HUMAN
|
||||||
| NC score | 0.042051 (rank : 28) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQ76 | Gene names | MEPE | |||
|
Domain Architecture |

|
|||||
| Description | Matrix extracellular phosphoglycoprotein precursor (Osteoblast/osteocyte factor 45) (OF45). | |||||
|
PAPPA_MOUSE
|
||||||
| NC score | 0.040676 (rank : 29) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R4K8, Q80VW3, Q80YY1, Q8K423, Q8R4K7, Q9ES06, Q9JK57 | Gene names | Pappa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.040299 (rank : 30) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.038485 (rank : 31) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
DPOLZ_HUMAN
|
||||||
| NC score | 0.036845 (rank : 32) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.035557 (rank : 33) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
JARD2_HUMAN
|
||||||
| NC score | 0.035166 (rank : 34) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
PAPPA_HUMAN
|
||||||
| NC score | 0.034754 (rank : 35) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
DPOLZ_MOUSE
|
||||||
| NC score | 0.033761 (rank : 36) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.033155 (rank : 37) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.032475 (rank : 38) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
X3CL1_MOUSE
|
||||||
| NC score | 0.032166 (rank : 39) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35188, O35933 | Gene names | Cx3cl1, Cx3c, Fkn, Scyd1 | |||
|
Domain Architecture |

|
|||||
| Description | Fractalkine precursor (CX3CL1) (Neurotactin) (CX3C membrane-anchored chemokine) (Small inducible cytokine D1). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.032038 (rank : 40) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
SEMG2_HUMAN
|
||||||
| NC score | 0.031971 (rank : 41) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
WDR20_HUMAN
|
||||||
| NC score | 0.029413 (rank : 42) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TBZ3, Q8WXX2, Q9UF86 | Gene names | WDR20 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 20 (DMR protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.028884 (rank : 43) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CN093_HUMAN
|
||||||
| NC score | 0.028497 (rank : 44) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H972, Q86SE6, Q96CF7, Q9HA68 | Gene names | C14orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf93 precursor. | |||||
|
SMBP2_MOUSE
|
||||||
| NC score | 0.028419 (rank : 45) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
BCLF1_HUMAN
|
||||||
| NC score | 0.027048 (rank : 46) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.026996 (rank : 47) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.026085 (rank : 48) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.025661 (rank : 49) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.023069 (rank : 50) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.021983 (rank : 51) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.021343 (rank : 52) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
ZCHC7_HUMAN
|
||||||
| NC score | 0.019474 (rank : 53) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N3Z6, Q5T0Q8, Q5T0Q9, Q5T0R0, Q8N2M1, Q8N4J2, Q8TBK8, Q9H648, Q9P0F0 | Gene names | ZCCHC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 7. | |||||
|
CENPT_HUMAN
|
||||||
| NC score | 0.019049 (rank : 54) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96BT3, Q96I29, Q96IC6, Q96NK9, Q9H901 | Gene names | CENPT, C16orf56, ICEN22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T) (Interphase centromere complex protein 22). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.017651 (rank : 55) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.017646 (rank : 56) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
COHA1_HUMAN
|
||||||
| NC score | 0.017071 (rank : 57) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
RBP2_MOUSE
|
||||||
| NC score | 0.016020 (rank : 58) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
TEX2_MOUSE
|
||||||
| NC score | 0.014493 (rank : 59) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZPJ0 | Gene names | Tex2, Kiaa1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
PROM1_HUMAN
|
||||||
| NC score | 0.014210 (rank : 60) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43490 | Gene names | PROM1, PROML1 | |||
|
Domain Architecture |

|
|||||
| Description | Prominin-1 precursor (Prominin-like protein 1) (Antigen AC133) (CD133 antigen). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.013148 (rank : 61) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
PRS23_HUMAN
|
||||||
| NC score | 0.011814 (rank : 62) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95084 | Gene names | PRSS23, ZSIG13 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease 23 precursor (EC 3.4.21.-) (Putative secreted protein ZSIG13). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.011507 (rank : 63) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
GRP75_MOUSE
|
||||||
| NC score | 0.011183 (rank : 64) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
GRP75_HUMAN
|
||||||
| NC score | 0.011148 (rank : 65) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
RNF6_HUMAN
|
||||||
| NC score | 0.011107 (rank : 66) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y252, Q9UF41 | Gene names | RNF6 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 6 (RING-H2 protein). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.010812 (rank : 67) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.009836 (rank : 68) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
ZN292_HUMAN
|
||||||
| NC score | 0.009597 (rank : 69) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
SIN3A_MOUSE
|
||||||
| NC score | 0.009519 (rank : 70) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
MAML1_MOUSE
|
||||||
| NC score | 0.009370 (rank : 71) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6T264, Q505D8, Q5SUC2, Q6PDK3, Q6ZQG5, Q8BIU5, Q8R3T0 | Gene names | Maml1, Kiaa0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.009236 (rank : 72) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
LUZP1_HUMAN
|
||||||
| NC score | 0.008538 (rank : 73) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.007886 (rank : 74) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
JADE3_MOUSE
|
||||||
| NC score | 0.007494 (rank : 75) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
ATS12_HUMAN
|
||||||
| NC score | 0.007320 (rank : 76) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58397, Q6UWL3 | Gene names | ADAMTS12 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
LIRA3_HUMAN
|
||||||
| NC score | 0.007026 (rank : 77) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6C8, O15469, O15470, O75016, Q8N151, Q8N154, Q8NHJ1, Q8NHJ2, Q8NHJ3, Q8NHJ4 | Gene names | LILRA3, ILT6, LIR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 3 precursor (Leukocyte immunoglobulin-like receptor 4) (LIR-4) (Immunoglobulin- like transcript 6) (ILT-6) (Monocyte inhibitory receptor HM43/HM31) (CD85e antigen). | |||||
|
ARD1_MOUSE
|
||||||
| NC score | 0.006816 (rank : 78) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGX0, Q8C2B6, Q8CDA4, Q8CDA7 | Gene names | Trim23, Arfd1 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein ARD-1 (ADP-ribosylation factor domain protein 1) (Tripartite motif-containing protein 23). | |||||
|
ARD1_HUMAN
|
||||||
| NC score | 0.006572 (rank : 79) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 621 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36406, Q9BZY4, Q9BZY5 | Gene names | TRIM23, ARD1, ARFD1, RNF46 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein ARD-1 (ADP-ribosylation factor domain protein 1) (Tripartite motif-containing protein 23) (RING finger protein 46). | |||||
|
ARHGC_HUMAN
|
||||||
| NC score | 0.006245 (rank : 80) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
K1196_HUMAN
|
||||||
| NC score | 0.005850 (rank : 81) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96KM6, Q9ULM4 | Gene names | KIAA1196 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1196. | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.005141 (rank : 82) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
CDKL5_HUMAN
|
||||||
| NC score | 0.005011 (rank : 83) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.004364 (rank : 84) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
MTR1L_HUMAN
|
||||||
| NC score | 0.001455 (rank : 85) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
EVI1_HUMAN
|
||||||
| NC score | 0.000582 (rank : 86) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
PLK4_HUMAN
|
||||||
| NC score | -0.000678 (rank : 87) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00444, Q8IYF0, Q96Q95 | Gene names | PLK4, SAK, STK18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PLK4 (EC 2.7.11.21) (Polo-like kinase 4) (PLK-4) (Serine/threonine-protein kinase Sak) (Serine/threonine- protein kinase 18). | |||||
|
ZN148_HUMAN
|
||||||
| NC score | -0.001122 (rank : 88) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UQR1, O00389, O43591 | Gene names | ZNF148, ZBP89 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89). | |||||