Please be patient as the page loads
|
CCNK_HUMAN
|
||||||
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CCNK_HUMAN
|
||||||
| θ value | 3.37801e-156 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 1.28364e-155 (rank : 2) | NC score | 0.857299 (rank : 2) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CCNT1_HUMAN
|
||||||
| θ value | 2.96089e-35 (rank : 3) | NC score | 0.814114 (rank : 4) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60563, O60581 | Gene names | CCNT1 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT1_MOUSE
|
||||||
| θ value | 2.96089e-35 (rank : 4) | NC score | 0.812881 (rank : 5) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT2_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 5) | NC score | 0.821365 (rank : 3) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
CCNL1_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 6) | NC score | 0.780984 (rank : 8) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CCNL1_MOUSE
|
||||||
| θ value | 4.28545e-26 (rank : 7) | NC score | 0.781011 (rank : 7) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
CCNL2_MOUSE
|
||||||
| θ value | 5.59698e-26 (rank : 8) | NC score | 0.789228 (rank : 6) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
CCNL2_HUMAN
|
||||||
| θ value | 1.24688e-25 (rank : 9) | NC score | 0.778885 (rank : 9) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
CCNC_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 10) | NC score | 0.703539 (rank : 10) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P24863, Q9H543 | Gene names | CCNC | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-C. | |||||
|
CCNC_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 11) | NC score | 0.685466 (rank : 11) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62447, Q9WUZ4 | Gene names | Ccnc | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-C. | |||||
|
CCNH_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 12) | NC score | 0.385641 (rank : 12) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51946, Q8TBL9 | Gene names | CCNH | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-H (MO15-associated protein) (p37) (p34). | |||||
|
CCND1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.188097 (rank : 17) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P25322 | Gene names | Ccnd1, Cyl-1 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D1. | |||||
|
CCNA2_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 14) | NC score | 0.187190 (rank : 18) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNH_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 15) | NC score | 0.354660 (rank : 13) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61458, Q9CVJ0, Q9JHV7 | Gene names | Ccnh | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-H. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 16) | NC score | 0.075365 (rank : 39) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CCNA1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 17) | NC score | 0.194757 (rank : 15) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 18) | NC score | 0.190888 (rank : 16) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-A1. | |||||
|
CCND1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 19) | NC score | 0.180751 (rank : 19) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P24385, Q6LEF0 | Gene names | CCND1, BCL1, PRAD1 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D1 (PRAD1 oncogene) (BCL-1 oncogene). | |||||
|
CCNA2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 20) | NC score | 0.201732 (rank : 14) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCND2_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 21) | NC score | 0.180251 (rank : 20) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P30280 | Gene names | Ccnd2, Cyl-2 | |||
|
Domain Architecture |
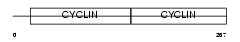
|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCND2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.174164 (rank : 21) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P30279, Q13955 | Gene names | CCND2 | |||
|
Domain Architecture |
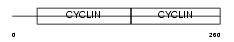
|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
SMG7_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.082580 (rank : 34) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.077993 (rank : 38) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
SMG7_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.079212 (rank : 37) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
ETV6_MOUSE
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.041712 (rank : 66) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.043663 (rank : 64) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.057309 (rank : 51) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.037438 (rank : 75) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CCNI_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.111056 (rank : 25) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
RTEL1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.033847 (rank : 84) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZ71, Q5JTV4, Q9BW37, Q9H402, Q9H4X6, Q9NX25, Q9NZ73, Q9UPR4, Q9Y4R6 | Gene names | RTEL1, C20orf41, KIAA1088, NHL | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of telomere elongation helicase 1 (EC 3.6.1.-) (Helicase- like protein NHL). | |||||
|
CCNI_MOUSE
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.104949 (rank : 26) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |
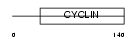
|
|||||
| Description | Cyclin-I. | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.035437 (rank : 82) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.036122 (rank : 80) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.061694 (rank : 47) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.051378 (rank : 55) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
STK10_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.001511 (rank : 119) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
TOX_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.039781 (rank : 71) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.122735 (rank : 22) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
GAK18_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.019062 (rank : 102) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.039364 (rank : 72) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.004707 (rank : 116) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.044245 (rank : 62) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.053022 (rank : 53) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.030868 (rank : 86) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ADA12_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.014452 (rank : 107) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
SYTL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.026354 (rank : 92) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
TOX_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.037433 (rank : 76) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
GLIS3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.002382 (rank : 118) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.026109 (rank : 93) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.037075 (rank : 77) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.055815 (rank : 52) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.050542 (rank : 57) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ETV6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.044100 (rank : 63) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.040091 (rank : 68) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.015481 (rank : 104) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.020898 (rank : 99) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
ZBT26_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.000205 (rank : 121) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HCK0, Q8WTR1 | Gene names | ZBTB26, KIAA1572, ZNF481 | |||
|
Domain Architecture |
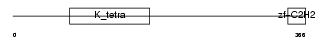
|
|||||
| Description | Zinc finger and BTB domain-containing protein 26 (Zinc finger protein 481) (Zinc finger protein Bioref). | |||||
|
K1210_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.038003 (rank : 74) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.015205 (rank : 105) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.007217 (rank : 112) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
PRGC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.046875 (rank : 60) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.047033 (rank : 59) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.065712 (rank : 46) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.050516 (rank : 58) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.020616 (rank : 100) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.044969 (rank : 61) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.035569 (rank : 81) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
ETV5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.025301 (rank : 94) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
F126A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.028811 (rank : 88) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P9N1, Q99NC4 | Gene names | Fam126a, Drctnnb1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM126A (Down-regulated by CTNNB1 protein A). | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.005709 (rank : 115) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.007924 (rank : 110) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.040034 (rank : 69) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.007373 (rank : 111) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.034935 (rank : 83) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.000438 (rank : 120) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.022960 (rank : 97) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.061118 (rank : 48) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NFIP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.036677 (rank : 78) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NV92, Q7Z2H3, Q7Z428, Q8TAR3, Q9ULQ5 | Gene names | NDFIP2, KIAA1165, N4WBP5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD4 family-interacting protein 2 (NEDD4 WW domain-binding protein 5A) (Putative MAPK-activating protein PM04/PM05/PM06/PM07) (Putative NF-kappa-B-activating protein 413). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.043556 (rank : 65) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.038111 (rank : 73) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.032439 (rank : 85) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.040233 (rank : 67) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
GA2L3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.026420 (rank : 90) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
NFRKB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.022122 (rank : 98) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.052332 (rank : 54) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.028250 (rank : 89) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PTPRG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.005942 (rank : 114) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
SOX13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.026405 (rank : 91) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
ZN438_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.013479 (rank : 108) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.030644 (rank : 87) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DAB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.014501 (rank : 106) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
FOXN4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.003912 (rank : 117) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K3Q3, Q920C0 | Gene names | Foxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N4. | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.013179 (rank : 109) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.040014 (rank : 70) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.006428 (rank : 113) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.036154 (rank : 79) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.016990 (rank : 103) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.019490 (rank : 101) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.023972 (rank : 96) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.024906 (rank : 95) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.073755 (rank : 40) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CCNB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.104611 (rank : 27) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P14635 | Gene names | CCNB1, CCNB | |||
|
Domain Architecture |
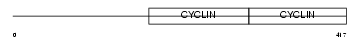
|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.103151 (rank : 28) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
Domain Architecture |
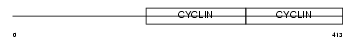
|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.086641 (rank : 29) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95067 | Gene names | CCNB2 | |||
|
Domain Architecture |
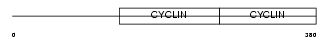
|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.085153 (rank : 30) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P30276 | Gene names | Ccnb2, Cycb2 | |||
|
Domain Architecture |
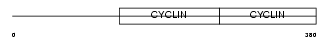
|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.079369 (rank : 36) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCND3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.116248 (rank : 23) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P30281, Q5T8J0, Q96F49 | Gene names | CCND3 | |||
|
Domain Architecture |
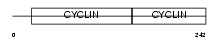
|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCND3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.116095 (rank : 24) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P30282, Q62391, Q99KP2 | Gene names | Ccnd3, Cyl-3 | |||
|
Domain Architecture |
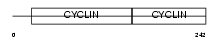
|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCNE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.083749 (rank : 32) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P24864, Q14091, Q8NFG1, Q92501 | Gene names | CCNE1, CCNE | |||
|
Domain Architecture |
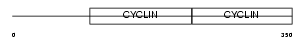
|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.082538 (rank : 35) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61457 | Gene names | Ccne1, Ccne | |||
|
Domain Architecture |
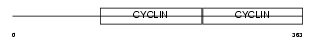
|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.084068 (rank : 31) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O96020, O95439 | Gene names | CCNE2 | |||
|
Domain Architecture |
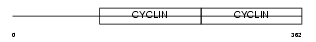
|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.083053 (rank : 33) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z238, Q9QWU5 | Gene names | Ccne2 | |||
|
Domain Architecture |
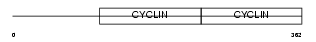
|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.072124 (rank : 41) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P41002, Q96EG9 | Gene names | CCNF | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.070780 (rank : 42) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.068020 (rank : 45) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51959, Q15757, Q96L32 | Gene names | CCNG1, CCNG, CYCG1 | |||
|
Domain Architecture |
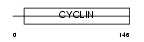
|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.070017 (rank : 43) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51945, O54779 | Gene names | Ccng1, Ccng | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.058678 (rank : 50) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |
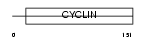
|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.060213 (rank : 49) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08918, O35612 | Gene names | Ccng2 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-G2. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.051266 (rank : 56) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.069556 (rank : 44) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CCNK_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.37801e-156 (rank : 1) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.857299 (rank : 2) | θ value | 1.28364e-155 (rank : 2) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CCNT2_HUMAN
|
||||||
| NC score | 0.821365 (rank : 3) | θ value | 1.52067e-31 (rank : 5) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
CCNT1_HUMAN
|
||||||
| NC score | 0.814114 (rank : 4) | θ value | 2.96089e-35 (rank : 3) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60563, O60581 | Gene names | CCNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT1_MOUSE
|
||||||
| NC score | 0.812881 (rank : 5) | θ value | 2.96089e-35 (rank : 4) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNL2_MOUSE
|
||||||
| NC score | 0.789228 (rank : 6) | θ value | 5.59698e-26 (rank : 8) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
CCNL1_MOUSE
|
||||||
| NC score | 0.781011 (rank : 7) | θ value | 4.28545e-26 (rank : 7) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
CCNL1_HUMAN
|
||||||
| NC score | 0.780984 (rank : 8) | θ value | 4.28545e-26 (rank : 6) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CCNL2_HUMAN
|
||||||
| NC score | 0.778885 (rank : 9) | θ value | 1.24688e-25 (rank : 9) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
CCNC_HUMAN
|
||||||
| NC score | 0.703539 (rank : 10) | θ value | 1.68911e-14 (rank : 10) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P24863, Q9H543 | Gene names | CCNC | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-C. | |||||
|
CCNC_MOUSE
|
||||||
| NC score | 0.685466 (rank : 11) | θ value | 4.16044e-13 (rank : 11) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62447, Q9WUZ4 | Gene names | Ccnc | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-C. | |||||
|
CCNH_HUMAN
|
||||||
| NC score | 0.385641 (rank : 12) | θ value | 0.00035302 (rank : 12) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51946, Q8TBL9 | Gene names | CCNH | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-H (MO15-associated protein) (p37) (p34). | |||||
|
CCNH_MOUSE
|
||||||
| NC score | 0.354660 (rank : 13) | θ value | 0.00175202 (rank : 15) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61458, Q9CVJ0, Q9JHV7 | Gene names | Ccnh | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-H. | |||||
|
CCNA2_HUMAN
|
||||||
| NC score | 0.201732 (rank : 14) | θ value | 0.00665767 (rank : 20) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA1_HUMAN
|
||||||
| NC score | 0.194757 (rank : 15) | θ value | 0.00509761 (rank : 17) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA1_MOUSE
|
||||||
| NC score | 0.190888 (rank : 16) | θ value | 0.00509761 (rank : 18) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCND1_MOUSE
|
||||||
| NC score | 0.188097 (rank : 17) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P25322 | Gene names | Ccnd1, Cyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D1. | |||||
|
CCNA2_MOUSE
|
||||||
| NC score | 0.187190 (rank : 18) | θ value | 0.00134147 (rank : 14) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCND1_HUMAN
|
||||||
| NC score | 0.180751 (rank : 19) | θ value | 0.00509761 (rank : 19) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P24385, Q6LEF0 | Gene names | CCND1, BCL1, PRAD1 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D1 (PRAD1 oncogene) (BCL-1 oncogene). | |||||
|
CCND2_MOUSE
|
||||||
| NC score | 0.180251 (rank : 20) | θ value | 0.00665767 (rank : 21) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P30280 | Gene names | Ccnd2, Cyl-2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCND2_HUMAN
|
||||||
| NC score | 0.174164 (rank : 21) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P30279, Q13955 | Gene names | CCND2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.122735 (rank : 22) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCND3_HUMAN
|
||||||
| NC score | 0.116248 (rank : 23) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P30281, Q5T8J0, Q96F49 | Gene names | CCND3 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCND3_MOUSE
|
||||||
| NC score | 0.116095 (rank : 24) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P30282, Q62391, Q99KP2 | Gene names | Ccnd3, Cyl-3 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCNI_HUMAN
|
||||||
| NC score | 0.111056 (rank : 25) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNI_MOUSE
|
||||||
| NC score | 0.104949 (rank : 26) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNB1_HUMAN
|
||||||
| NC score | 0.104611 (rank : 27) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P14635 | Gene names | CCNB1, CCNB | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB1_MOUSE
|
||||||
| NC score | 0.103151 (rank : 28) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB2_HUMAN
|
||||||
| NC score | 0.086641 (rank : 29) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95067 | Gene names | CCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB2_MOUSE
|
||||||
| NC score | 0.085153 (rank : 30) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P30276 | Gene names | Ccnb2, Cycb2 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNE2_HUMAN
|
||||||
| NC score | 0.084068 (rank : 31) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O96020, O95439 | Gene names | CCNE2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE1_HUMAN
|
||||||
| NC score | 0.083749 (rank : 32) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P24864, Q14091, Q8NFG1, Q92501 | Gene names | CCNE1, CCNE | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNE2_MOUSE
|
||||||
| NC score | 0.083053 (rank : 33) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z238, Q9QWU5 | Gene names | Ccne2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
SMG7_MOUSE
|
||||||
| NC score | 0.082580 (rank : 34) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
CCNE1_MOUSE
|
||||||
| NC score | 0.082538 (rank : 35) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61457 | Gene names | Ccne1, Ccne | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.079369 (rank : 36) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
SMG7_HUMAN
|
||||||
| NC score | 0.079212 (rank : 37) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.077993 (rank : 38) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.075365 (rank : 39) | θ value | 0.00390308 (rank : 16) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.073755 (rank : 40) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CCNF_HUMAN
|
||||||
| NC score | 0.072124 (rank : 41) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P41002, Q96EG9 | Gene names | CCNF | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNF_MOUSE
|
||||||
| NC score | 0.070780 (rank : 42) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNG1_MOUSE
|
||||||
| NC score | 0.070017 (rank : 43) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51945, O54779 | Gene names | Ccng1, Ccng | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.069556 (rank : 44) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CCNG1_HUMAN
|
||||||
| NC score | 0.068020 (rank : 45) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51959, Q15757, Q96L32 | Gene names | CCNG1, CCNG, CYCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.065712 (rank : 46) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.061694 (rank : 47) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.061118 (rank : 48) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
CCNG2_MOUSE
|
||||||
| NC score | 0.060213 (rank : 49) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08918, O35612 | Gene names | Ccng2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG2_HUMAN
|
||||||
| NC score | 0.058678 (rank : 50) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.057309 (rank : 51) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.055815 (rank : 52) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.053022 (rank : 53) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.052332 (rank : 54) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.051378 (rank : 55) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.051266 (rank : 56) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.050542 (rank : 57) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.050516 (rank : 58) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.047033 (rank : 59) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
PRGC2_MOUSE
|
||||||
| NC score | 0.046875 (rank : 60) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.044969 (rank : 61) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.044245 (rank : 62) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
ETV6_HUMAN
|
||||||
| NC score | 0.044100 (rank : 63) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.043663 (rank : 64) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.043556 (rank : 65) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ETV6_MOUSE
|
||||||
| NC score | 0.041712 (rank : 66) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.040233 (rank : 67) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.040091 (rank : 68) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.040034 (rank : 69) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.040014 (rank : 70) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TOX_MOUSE
|
||||||
| NC score | 0.039781 (rank : 71) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.039364 (rank : 72) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.038111 (rank : 73) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
K1210_HUMAN
|
||||||
| NC score | 0.038003 (rank : 74) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.037438 (rank : 75) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
TOX_HUMAN
|
||||||
| NC score | 0.037433 (rank : 76) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.037075 (rank : 77) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFIP2_HUMAN
|
||||||
| NC score | 0.036677 (rank : 78) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NV92, Q7Z2H3, Q7Z428, Q8TAR3, Q9ULQ5 | Gene names | NDFIP2, KIAA1165, N4WBP5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD4 family-interacting protein 2 (NEDD4 WW domain-binding protein 5A) (Putative MAPK-activating protein PM04/PM05/PM06/PM07) (Putative NF-kappa-B-activating protein 413). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.036154 (rank : 79) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.036122 (rank : 80) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.035569 (rank : 81) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.035437 (rank : 82) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.034935 (rank : 83) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
RTEL1_HUMAN
|
||||||
| NC score | 0.033847 (rank : 84) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZ71, Q5JTV4, Q9BW37, Q9H402, Q9H4X6, Q9NX25, Q9NZ73, Q9UPR4, Q9Y4R6 | Gene names | RTEL1, C20orf41, KIAA1088, NHL | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of telomere elongation helicase 1 (EC 3.6.1.-) (Helicase- like protein NHL). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.032439 (rank : 85) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.030868 (rank : 86) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.030644 (rank : 87) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
F126A_MOUSE
|
||||||
| NC score | 0.028811 (rank : 88) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P9N1, Q99NC4 | Gene names | Fam126a, Drctnnb1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM126A (Down-regulated by CTNNB1 protein A). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.028250 (rank : 89) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
GA2L3_HUMAN
|
||||||
| NC score | 0.026420 (rank : 90) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
SOX13_HUMAN
|
||||||
| NC score | 0.026405 (rank : 91) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
SYTL1_HUMAN
|
||||||
| NC score | 0.026354 (rank : 92) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.026109 (rank : 93) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.025301 (rank : 94) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.024906 (rank : 95) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.023972 (rank : 96) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.022960 (rank : 97) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
NFRKB_MOUSE
|
||||||
| NC score | 0.022122 (rank : 98) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.020898 (rank : 99) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.020616 (rank : 100) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.019490 (rank : 101) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
GAK18_HUMAN
|
||||||
| NC score | 0.019062 (rank : 102) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.016990 (rank : 103) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.015481 (rank : 104) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.015205 (rank : 105) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
DAB1_HUMAN
|
||||||
| NC score | 0.014501 (rank : 106) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
ADA12_MOUSE
|
||||||
| NC score | 0.014452 (rank : 107) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
ZN438_HUMAN
|
||||||
| NC score | 0.013479 (rank : 108) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.013179 (rank : 109) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.007924 (rank : 110) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.007373 (rank : 111) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.007217 (rank : 112) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.006428 (rank : 113) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PTPRG_HUMAN
|
||||||
| NC score | 0.005942 (rank : 114) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
LATS1_MOUSE
|
||||||
| NC score | 0.005709 (rank : 115) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.004707 (rank : 116) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
FOXN4_MOUSE
|
||||||
| NC score | 0.003912 (rank : 117) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K3Q3, Q920C0 | Gene names | Foxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N4. | |||||
|
GLIS3_MOUSE
|
||||||
| NC score | 0.002382 (rank : 118) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.001511 (rank : 119) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | 0.000438 (rank : 120) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
ZBT26_HUMAN
|
||||||
| NC score | 0.000205 (rank : 121) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HCK0, Q8WTR1 | Gene names | ZBTB26, KIAA1572, ZNF481 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 26 (Zinc finger protein 481) (Zinc finger protein Bioref). | |||||