Please be patient as the page loads
|
CCNB1_HUMAN
|
||||||
| SwissProt Accessions | P14635 | Gene names | CCNB1, CCNB | |||
|
Domain Architecture |
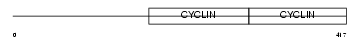
|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CCNB1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P14635 | Gene names | CCNB1, CCNB | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995640 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
Domain Architecture |
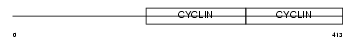
|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB2_HUMAN
|
||||||
| θ value | 4.04623e-109 (rank : 3) | NC score | 0.981667 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95067 | Gene names | CCNB2 | |||
|
Domain Architecture |
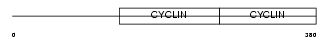
|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB2_MOUSE
|
||||||
| θ value | 5.46864e-106 (rank : 4) | NC score | 0.980647 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30276 | Gene names | Ccnb2, Cycb2 | |||
|
Domain Architecture |
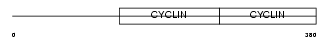
|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 1.32601e-43 (rank : 5) | NC score | 0.747789 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNA1_HUMAN
|
||||||
| θ value | 6.58091e-43 (rank : 6) | NC score | 0.914784 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA1_MOUSE
|
||||||
| θ value | 7.27602e-42 (rank : 7) | NC score | 0.914696 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-A1. | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 1.62093e-41 (rank : 8) | NC score | 0.840034 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNA2_MOUSE
|
||||||
| θ value | 4.71619e-41 (rank : 9) | NC score | 0.905794 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA2_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 10) | NC score | 0.903693 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNE1_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 11) | NC score | 0.793348 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P24864, Q14091, Q8NFG1, Q92501 | Gene names | CCNE1, CCNE | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNE2_MOUSE
|
||||||
| θ value | 4.15078e-21 (rank : 12) | NC score | 0.797328 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z238, Q9QWU5 | Gene names | Ccne2 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE1_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 13) | NC score | 0.784473 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61457 | Gene names | Ccne1, Ccne | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNE2_HUMAN
|
||||||
| θ value | 5.99374e-20 (rank : 14) | NC score | 0.791630 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O96020, O95439 | Gene names | CCNE2 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNF_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 15) | NC score | 0.705529 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P41002, Q96EG9 | Gene names | CCNF | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCND1_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 16) | NC score | 0.744344 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P25322 | Gene names | Ccnd1, Cyl-1 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D1. | |||||
|
CCNF_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 17) | NC score | 0.702533 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCND1_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 18) | NC score | 0.736150 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P24385, Q6LEF0 | Gene names | CCND1, BCL1, PRAD1 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D1 (PRAD1 oncogene) (BCL-1 oncogene). | |||||
|
CCND3_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 19) | NC score | 0.722386 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30281, Q5T8J0, Q96F49 | Gene names | CCND3 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCND3_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 20) | NC score | 0.711437 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30282, Q62391, Q99KP2 | Gene names | Ccnd3, Cyl-3 | |||
|
Domain Architecture |
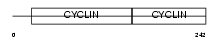
|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCND2_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 21) | NC score | 0.694114 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30280 | Gene names | Ccnd2, Cyl-2 | |||
|
Domain Architecture |
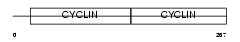
|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCND2_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 22) | NC score | 0.701282 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30279, Q13955 | Gene names | CCND2 | |||
|
Domain Architecture |
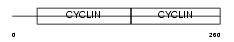
|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCNI_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 23) | NC score | 0.482433 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |
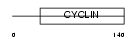
|
|||||
| Description | Cyclin-I. | |||||
|
CCNI_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 24) | NC score | 0.504835 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNG1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 25) | NC score | 0.454822 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51959, Q15757, Q96L32 | Gene names | CCNG1, CCNG, CYCG1 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 26) | NC score | 0.468017 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51945, O54779 | Gene names | Ccng1, Ccng | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 27) | NC score | 0.428164 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08918, O35612 | Gene names | Ccng2 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-G2. | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 28) | NC score | 0.044489 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
CCNG2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 29) | NC score | 0.425130 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-G2. | |||||
|
UNG2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 30) | NC score | 0.347358 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22674 | Gene names | UNG2 | |||
|
Domain Architecture |
|
|||||
| Description | Uracil-DNA glycosylase 2 (EC 3.2.2.-) (UDG 2). | |||||
|
CCNL2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.077182 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.013817 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
CCNL2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.068020 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.025649 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.020330 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
ZN462_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.002476 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JM2, Q8N408 | Gene names | ZNF462, KIAA1803 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 462. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.011320 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
GCNL2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.016159 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
M3K4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | -0.000027 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||
|
RM39_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.024729 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JKF7, Q91YK5, Q9D8U5 | Gene names | Mrpl39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 39S ribosomal protein L39 (L39mt) (MRP-L39). | |||||
|
GPR64_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.007022 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
PO6F1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.005806 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.002117 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SNX9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.009859 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5X1, Q9BSI7, Q9BVM1, Q9UJH6, Q9UP20 | Gene names | SNX9, SH3PX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-9 (SH3 and PX domain-containing protein 1) (SDP1 protein). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.018725 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
ECM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.013330 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.001689 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CCNH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.051169 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61458, Q9CVJ0, Q9JHV7 | Gene names | Ccnh | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-H. | |||||
|
CCNL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.068984 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CCNL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.068881 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.004484 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
NFM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.002474 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
RM39_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.020304 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYK5, Q5QTR3, Q96Q65, Q9BSQ7, Q9BZV6, Q9NX44 | Gene names | MRPL39, MRPL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 39S ribosomal protein L39 (L39mt) (MRP-L39) (MRP-L5). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.014832 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
GGPPS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.015342 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTN0 | Gene names | Ggps1 | |||
|
Domain Architecture |
|
|||||
| Description | Geranylgeranyl pyrophosphate synthetase (GGPP synthetase) (GGPPSase) (Geranylgeranyl diphosphate synthase) [Includes: Dimethylallyltranstransferase (EC 2.5.1.1); Geranyltranstransferase (EC 2.5.1.10); Farnesyltranstransferase (EC 2.5.1.29)]. | |||||
|
JHD3D_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.004069 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6B0I6, Q9NT41, Q9NW76 | Gene names | JMJD2D, JHDM3D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.029685 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.007672 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TACC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.002493 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
ZFYV9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.005071 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
ZHX3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.005157 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H4I2, O43145 | Gene names | ZHX3, KIAA0395, TIX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and homeodomain protein 3 (Triple homeobox protein 1). | |||||
|
CCNK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.104611 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.085787 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CCNB1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P14635 | Gene names | CCNB1, CCNB | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB1_MOUSE
|
||||||
| NC score | 0.995640 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB2_HUMAN
|
||||||
| NC score | 0.981667 (rank : 3) | θ value | 4.04623e-109 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95067 | Gene names | CCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB2_MOUSE
|
||||||
| NC score | 0.980647 (rank : 4) | θ value | 5.46864e-106 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30276 | Gene names | Ccnb2, Cycb2 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNA1_HUMAN
|
||||||
| NC score | 0.914784 (rank : 5) | θ value | 6.58091e-43 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA1_MOUSE
|
||||||
| NC score | 0.914696 (rank : 6) | θ value | 7.27602e-42 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA2_MOUSE
|
||||||
| NC score | 0.905794 (rank : 7) | θ value | 4.71619e-41 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA2_HUMAN
|
||||||
| NC score | 0.903693 (rank : 8) | θ value | 2.34062e-40 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.840034 (rank : 9) | θ value | 1.62093e-41 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNE2_MOUSE
|
||||||
| NC score | 0.797328 (rank : 10) | θ value | 4.15078e-21 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z238, Q9QWU5 | Gene names | Ccne2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE1_HUMAN
|
||||||
| NC score | 0.793348 (rank : 11) | θ value | 4.15078e-21 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P24864, Q14091, Q8NFG1, Q92501 | Gene names | CCNE1, CCNE | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNE2_HUMAN
|
||||||
| NC score | 0.791630 (rank : 12) | θ value | 5.99374e-20 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O96020, O95439 | Gene names | CCNE2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE1_MOUSE
|
||||||
| NC score | 0.784473 (rank : 13) | θ value | 2.06002e-20 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61457 | Gene names | Ccne1, Ccne | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.747789 (rank : 14) | θ value | 1.32601e-43 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCND1_MOUSE
|
||||||
| NC score | 0.744344 (rank : 15) | θ value | 6.20254e-17 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P25322 | Gene names | Ccnd1, Cyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D1. | |||||
|
CCND1_HUMAN
|
||||||
| NC score | 0.736150 (rank : 16) | θ value | 1.16975e-15 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P24385, Q6LEF0 | Gene names | CCND1, BCL1, PRAD1 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D1 (PRAD1 oncogene) (BCL-1 oncogene). | |||||
|
CCND3_HUMAN
|
||||||
| NC score | 0.722386 (rank : 17) | θ value | 1.29331e-14 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30281, Q5T8J0, Q96F49 | Gene names | CCND3 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCND3_MOUSE
|
||||||
| NC score | 0.711437 (rank : 18) | θ value | 2.43908e-13 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30282, Q62391, Q99KP2 | Gene names | Ccnd3, Cyl-3 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCNF_HUMAN
|
||||||
| NC score | 0.705529 (rank : 19) | θ value | 1.24977e-17 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P41002, Q96EG9 | Gene names | CCNF | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNF_MOUSE
|
||||||
| NC score | 0.702533 (rank : 20) | θ value | 1.80466e-16 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCND2_HUMAN
|
||||||
| NC score | 0.701282 (rank : 21) | θ value | 6.00763e-12 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30279, Q13955 | Gene names | CCND2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCND2_MOUSE
|
||||||
| NC score | 0.694114 (rank : 22) | θ value | 4.59992e-12 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30280 | Gene names | Ccnd2, Cyl-2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCNI_HUMAN
|
||||||
| NC score | 0.504835 (rank : 23) | θ value | 1.87187e-05 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNI_MOUSE
|
||||||
| NC score | 0.482433 (rank : 24) | θ value | 8.40245e-06 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNG1_MOUSE
|
||||||
| NC score | 0.468017 (rank : 25) | θ value | 0.00869519 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51945, O54779 | Gene names | Ccng1, Ccng | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG1_HUMAN
|
||||||
| NC score | 0.454822 (rank : 26) | θ value | 0.00869519 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51959, Q15757, Q96L32 | Gene names | CCNG1, CCNG, CYCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG2_MOUSE
|
||||||
| NC score | 0.428164 (rank : 27) | θ value | 0.00869519 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08918, O35612 | Gene names | Ccng2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG2_HUMAN
|
||||||
| NC score | 0.425130 (rank : 28) | θ value | 0.0193708 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
UNG2_HUMAN
|
||||||
| NC score | 0.347358 (rank : 29) | θ value | 0.0431538 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22674 | Gene names | UNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Uracil-DNA glycosylase 2 (EC 3.2.2.-) (UDG 2). | |||||
|
CCNK_HUMAN
|
||||||
| NC score | 0.104611 (rank : 30) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.085787 (rank : 31) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CCNL2_MOUSE
|
||||||
| NC score | 0.077182 (rank : 32) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
CCNL1_HUMAN
|
||||||
| NC score | 0.068984 (rank : 33) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CCNL1_MOUSE
|
||||||
| NC score | 0.068881 (rank : 34) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
CCNL2_HUMAN
|
||||||
| NC score | 0.068020 (rank : 35) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
CCNH_MOUSE
|
||||||
| NC score | 0.051169 (rank : 36) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61458, Q9CVJ0, Q9JHV7 | Gene names | Ccnh | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-H. | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.044489 (rank : 37) | θ value | 0.0113563 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.029685 (rank : 38) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.025649 (rank : 39) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
RM39_MOUSE
|
||||||
| NC score | 0.024729 (rank : 40) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JKF7, Q91YK5, Q9D8U5 | Gene names | Mrpl39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 39S ribosomal protein L39 (L39mt) (MRP-L39). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.020330 (rank : 41) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
RM39_HUMAN
|
||||||
| NC score | 0.020304 (rank : 42) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYK5, Q5QTR3, Q96Q65, Q9BSQ7, Q9BZV6, Q9NX44 | Gene names | MRPL39, MRPL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 39S ribosomal protein L39 (L39mt) (MRP-L39) (MRP-L5). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.018725 (rank : 43) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
GCNL2_MOUSE
|
||||||
| NC score | 0.016159 (rank : 44) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
GGPPS_MOUSE
|
||||||
| NC score | 0.015342 (rank : 45) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTN0 | Gene names | Ggps1 | |||
|
Domain Architecture |
|
|||||
| Description | Geranylgeranyl pyrophosphate synthetase (GGPP synthetase) (GGPPSase) (Geranylgeranyl diphosphate synthase) [Includes: Dimethylallyltranstransferase (EC 2.5.1.1); Geranyltranstransferase (EC 2.5.1.10); Farnesyltranstransferase (EC 2.5.1.29)]. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.014832 (rank : 46) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.013817 (rank : 47) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
ECM1_HUMAN
|
||||||
| NC score | 0.013330 (rank : 48) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.011320 (rank : 49) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
SNX9_HUMAN
|
||||||
| NC score | 0.009859 (rank : 50) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5X1, Q9BSI7, Q9BVM1, Q9UJH6, Q9UP20 | Gene names | SNX9, SH3PX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-9 (SH3 and PX domain-containing protein 1) (SDP1 protein). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.007672 (rank : 51) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
GPR64_HUMAN
|
||||||
| NC score | 0.007022 (rank : 52) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
PO6F1_MOUSE
|
||||||
| NC score | 0.005806 (rank : 53) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
ZHX3_HUMAN
|
||||||
| NC score | 0.005157 (rank : 54) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H4I2, O43145 | Gene names | ZHX3, KIAA0395, TIX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and homeodomain protein 3 (Triple homeobox protein 1). | |||||
|
ZFYV9_HUMAN
|
||||||
| NC score | 0.005071 (rank : 55) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.004484 (rank : 56) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
JHD3D_HUMAN
|
||||||
| NC score | 0.004069 (rank : 57) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6B0I6, Q9NT41, Q9NW76 | Gene names | JMJD2D, JHDM3D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
TACC1_MOUSE
|
||||||
| NC score | 0.002493 (rank : 58) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
ZN462_HUMAN
|
||||||
| NC score | 0.002476 (rank : 59) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JM2, Q8N408 | Gene names | ZNF462, KIAA1803 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 462. | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.002474 (rank : 60) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.002117 (rank : 61) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.001689 (rank : 62) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
M3K4_MOUSE
|
||||||
| NC score | -0.000027 (rank : 63) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||