Please be patient as the page loads
|
CCNA2_MOUSE
|
||||||
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CCNA2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995307 (rank : 2) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA1_HUMAN
|
||||||
| θ value | 3.55291e-97 (rank : 3) | NC score | 0.977244 (rank : 4) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA1_MOUSE
|
||||||
| θ value | 2.63489e-92 (rank : 4) | NC score | 0.979330 (rank : 3) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-A1. | |||||
|
CCNB1_HUMAN
|
||||||
| θ value | 4.71619e-41 (rank : 5) | NC score | 0.905794 (rank : 7) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P14635 | Gene names | CCNB1, CCNB | |||
|
Domain Architecture |
|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB1_MOUSE
|
||||||
| θ value | 1.05066e-40 (rank : 6) | NC score | 0.907365 (rank : 5) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
Domain Architecture |
|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB2_HUMAN
|
||||||
| θ value | 1.85459e-37 (rank : 7) | NC score | 0.906222 (rank : 6) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95067 | Gene names | CCNB2 | |||
|
Domain Architecture |
|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB2_MOUSE
|
||||||
| θ value | 7.04741e-37 (rank : 8) | NC score | 0.902826 (rank : 8) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30276 | Gene names | Ccnb2, Cycb2 | |||
|
Domain Architecture |
|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 1.73584e-35 (rank : 9) | NC score | 0.829687 (rank : 9) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 6.59618e-35 (rank : 10) | NC score | 0.723239 (rank : 22) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNF_HUMAN
|
||||||
| θ value | 3.07116e-24 (rank : 11) | NC score | 0.733728 (rank : 20) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P41002, Q96EG9 | Gene names | CCNF | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNE1_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 12) | NC score | 0.801012 (rank : 13) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61457 | Gene names | Ccne1, Ccne | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNF_MOUSE
|
||||||
| θ value | 1.6852e-22 (rank : 13) | NC score | 0.731710 (rank : 21) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNE1_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 14) | NC score | 0.806020 (rank : 10) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P24864, Q14091, Q8NFG1, Q92501 | Gene names | CCNE1, CCNE | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNE2_HUMAN
|
||||||
| θ value | 2.43343e-21 (rank : 15) | NC score | 0.805671 (rank : 11) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O96020, O95439 | Gene names | CCNE2 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE2_MOUSE
|
||||||
| θ value | 2.69047e-20 (rank : 16) | NC score | 0.805504 (rank : 12) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z238, Q9QWU5 | Gene names | Ccne2 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCND3_HUMAN
|
||||||
| θ value | 5.99374e-20 (rank : 17) | NC score | 0.776595 (rank : 16) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30281, Q5T8J0, Q96F49 | Gene names | CCND3 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCND1_HUMAN
|
||||||
| θ value | 1.74391e-19 (rank : 18) | NC score | 0.789410 (rank : 15) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P24385, Q6LEF0 | Gene names | CCND1, BCL1, PRAD1 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D1 (PRAD1 oncogene) (BCL-1 oncogene). | |||||
|
CCND1_MOUSE
|
||||||
| θ value | 2.27762e-19 (rank : 19) | NC score | 0.792456 (rank : 14) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P25322 | Gene names | Ccnd1, Cyl-1 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D1. | |||||
|
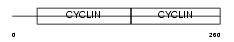
CCND2_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 20) | NC score | 0.765927 (rank : 17) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P30279, Q13955 | Gene names | CCND2 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
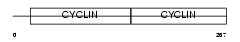
CCND2_MOUSE
|
||||||
| θ value | 1.47631e-18 (rank : 21) | NC score | 0.757501 (rank : 19) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P30280 | Gene names | Ccnd2, Cyl-2 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
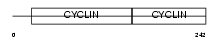
CCND3_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 22) | NC score | 0.762989 (rank : 18) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30282, Q62391, Q99KP2 | Gene names | Ccnd3, Cyl-3 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
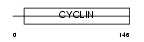
CCNG1_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 23) | NC score | 0.551811 (rank : 24) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51959, Q15757, Q96L32 | Gene names | CCNG1, CCNG, CYCG1 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG1_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 24) | NC score | 0.565943 (rank : 23) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51945, O54779 | Gene names | Ccng1, Ccng | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNI_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 25) | NC score | 0.541358 (rank : 25) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNG2_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 26) | NC score | 0.500766 (rank : 28) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG2_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 27) | NC score | 0.504016 (rank : 27) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08918, O35612 | Gene names | Ccng2 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-G2. | |||||
|
UNG2_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 28) | NC score | 0.393149 (rank : 29) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22674 | Gene names | UNG2 | |||
|
Domain Architecture |
|
|||||
| Description | Uracil-DNA glycosylase 2 (EC 3.2.2.-) (UDG 2). | |||||
|
CCNI_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 29) | NC score | 0.514389 (rank : 26) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-I. | |||||
|
CCNK_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 30) | NC score | 0.187190 (rank : 30) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 31) | NC score | 0.155589 (rank : 31) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
ZF106_MOUSE
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.021156 (rank : 43) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
CCNL1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.112078 (rank : 33) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CCNL1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.111864 (rank : 34) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
CCNL2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.112908 (rank : 32) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
CCNL2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.101940 (rank : 35) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.009681 (rank : 45) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.002521 (rank : 50) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
S27A4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.009062 (rank : 46) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VE0, O88562 | Gene names | Slc27a4, Acsvl4, Fatp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Long-chain fatty acid transport protein 4 (EC 6.2.1.-) (Fatty acid transport protein 4) (FATP-4) (Solute carrier family 27 member 4). | |||||
|
FHOD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.007370 (rank : 48) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y613, Q59F76, Q6Y1F2, Q76MS8, Q8N521 | Gene names | FHOD1, FHOS | |||
|
Domain Architecture |

|
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
PTRF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.007662 (rank : 47) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NZI2, O00535, Q6GMY1, Q96H74, Q9BT85, Q9HAP4 | Gene names | PTRF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polymerase I and transcript release factor (PTRF protein). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | -0.000547 (rank : 51) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.003734 (rank : 49) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MINT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.011856 (rank : 44) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CCNC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.061066 (rank : 36) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24863, Q9H543 | Gene names | CCNC | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-C. | |||||
|
CCNC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.053827 (rank : 42) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62447, Q9WUZ4 | Gene names | Ccnc | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-C. | |||||
|
CCNH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.056339 (rank : 40) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51946, Q8TBL9 | Gene names | CCNH | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-H (MO15-associated protein) (p37) (p34). | |||||
|
CCNH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.059231 (rank : 37) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61458, Q9CVJ0, Q9JHV7 | Gene names | Ccnh | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-H. | |||||
|
CCNT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.057263 (rank : 38) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60563, O60581 | Gene names | CCNT1 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.055371 (rank : 41) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.056445 (rank : 39) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
CCNA2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA2_HUMAN
|
||||||
| NC score | 0.995307 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA1_MOUSE
|
||||||
| NC score | 0.979330 (rank : 3) | θ value | 2.63489e-92 (rank : 4) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA1_HUMAN
|
||||||
| NC score | 0.977244 (rank : 4) | θ value | 3.55291e-97 (rank : 3) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNB1_MOUSE
|
||||||
| NC score | 0.907365 (rank : 5) | θ value | 1.05066e-40 (rank : 6) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB2_HUMAN
|
||||||
| NC score | 0.906222 (rank : 6) | θ value | 1.85459e-37 (rank : 7) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95067 | Gene names | CCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB1_HUMAN
|
||||||
| NC score | 0.905794 (rank : 7) | θ value | 4.71619e-41 (rank : 5) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P14635 | Gene names | CCNB1, CCNB | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB2_MOUSE
|
||||||
| NC score | 0.902826 (rank : 8) | θ value | 7.04741e-37 (rank : 8) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30276 | Gene names | Ccnb2, Cycb2 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.829687 (rank : 9) | θ value | 1.73584e-35 (rank : 9) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNE1_HUMAN
|
||||||
| NC score | 0.806020 (rank : 10) | θ value | 2.87452e-22 (rank : 14) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P24864, Q14091, Q8NFG1, Q92501 | Gene names | CCNE1, CCNE | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNE2_HUMAN
|
||||||
| NC score | 0.805671 (rank : 11) | θ value | 2.43343e-21 (rank : 15) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O96020, O95439 | Gene names | CCNE2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE2_MOUSE
|
||||||
| NC score | 0.805504 (rank : 12) | θ value | 2.69047e-20 (rank : 16) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z238, Q9QWU5 | Gene names | Ccne2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE1_MOUSE
|
||||||
| NC score | 0.801012 (rank : 13) | θ value | 4.43474e-23 (rank : 12) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61457 | Gene names | Ccne1, Ccne | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCND1_MOUSE
|
||||||
| NC score | 0.792456 (rank : 14) | θ value | 2.27762e-19 (rank : 19) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P25322 | Gene names | Ccnd1, Cyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D1. | |||||
|
CCND1_HUMAN
|
||||||
| NC score | 0.789410 (rank : 15) | θ value | 1.74391e-19 (rank : 18) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P24385, Q6LEF0 | Gene names | CCND1, BCL1, PRAD1 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D1 (PRAD1 oncogene) (BCL-1 oncogene). | |||||
|
CCND3_HUMAN
|
||||||
| NC score | 0.776595 (rank : 16) | θ value | 5.99374e-20 (rank : 17) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30281, Q5T8J0, Q96F49 | Gene names | CCND3 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCND2_HUMAN
|
||||||
| NC score | 0.765927 (rank : 17) | θ value | 5.07402e-19 (rank : 20) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P30279, Q13955 | Gene names | CCND2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCND3_MOUSE
|
||||||
| NC score | 0.762989 (rank : 18) | θ value | 2.5182e-18 (rank : 22) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30282, Q62391, Q99KP2 | Gene names | Ccnd3, Cyl-3 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCND2_MOUSE
|
||||||
| NC score | 0.757501 (rank : 19) | θ value | 1.47631e-18 (rank : 21) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P30280 | Gene names | Ccnd2, Cyl-2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCNF_HUMAN
|
||||||
| NC score | 0.733728 (rank : 20) | θ value | 3.07116e-24 (rank : 11) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P41002, Q96EG9 | Gene names | CCNF | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNF_MOUSE
|
||||||
| NC score | 0.731710 (rank : 21) | θ value | 1.6852e-22 (rank : 13) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.723239 (rank : 22) | θ value | 6.59618e-35 (rank : 10) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNG1_MOUSE
|
||||||
| NC score | 0.565943 (rank : 23) | θ value | 5.62301e-10 (rank : 24) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51945, O54779 | Gene names | Ccng1, Ccng | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG1_HUMAN
|
||||||
| NC score | 0.551811 (rank : 24) | θ value | 3.29651e-10 (rank : 23) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51959, Q15757, Q96L32 | Gene names | CCNG1, CCNG, CYCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNI_HUMAN
|
||||||
| NC score | 0.541358 (rank : 25) | θ value | 6.43352e-06 (rank : 25) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNI_MOUSE
|
||||||
| NC score | 0.514389 (rank : 26) | θ value | 4.1701e-05 (rank : 29) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNG2_MOUSE
|
||||||
| NC score | 0.504016 (rank : 27) | θ value | 1.43324e-05 (rank : 27) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08918, O35612 | Gene names | Ccng2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG2_HUMAN
|
||||||
| NC score | 0.500766 (rank : 28) | θ value | 8.40245e-06 (rank : 26) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
UNG2_HUMAN
|
||||||
| NC score | 0.393149 (rank : 29) | θ value | 1.87187e-05 (rank : 28) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22674 | Gene names | UNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Uracil-DNA glycosylase 2 (EC 3.2.2.-) (UDG 2). | |||||
|
CCNK_HUMAN
|
||||||
| NC score | 0.187190 (rank : 30) | θ value | 0.00134147 (rank : 30) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.155589 (rank : 31) | θ value | 0.00228821 (rank : 31) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CCNL2_MOUSE
|
||||||
| NC score | 0.112908 (rank : 32) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
CCNL1_HUMAN
|
||||||
| NC score | 0.112078 (rank : 33) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CCNL1_MOUSE
|
||||||
| NC score | 0.111864 (rank : 34) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
CCNL2_HUMAN
|
||||||
| NC score | 0.101940 (rank : 35) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
CCNC_HUMAN
|
||||||
| NC score | 0.061066 (rank : 36) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24863, Q9H543 | Gene names | CCNC | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-C. | |||||
|
CCNH_MOUSE
|
||||||
| NC score | 0.059231 (rank : 37) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61458, Q9CVJ0, Q9JHV7 | Gene names | Ccnh | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-H. | |||||
|
CCNT1_HUMAN
|
||||||
| NC score | 0.057263 (rank : 38) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60563, O60581 | Gene names | CCNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT2_HUMAN
|
||||||
| NC score | 0.056445 (rank : 39) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
CCNH_HUMAN
|
||||||
| NC score | 0.056339 (rank : 40) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51946, Q8TBL9 | Gene names | CCNH | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-H (MO15-associated protein) (p37) (p34). | |||||
|
CCNT1_MOUSE
|
||||||
| NC score | 0.055371 (rank : 41) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNC_MOUSE
|
||||||
| NC score | 0.053827 (rank : 42) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62447, Q9WUZ4 | Gene names | Ccnc | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-C. | |||||
|
ZF106_MOUSE
|
||||||
| NC score | 0.021156 (rank : 43) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.011856 (rank : 44) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.009681 (rank : 45) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
S27A4_MOUSE
|
||||||
| NC score | 0.009062 (rank : 46) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VE0, O88562 | Gene names | Slc27a4, Acsvl4, Fatp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Long-chain fatty acid transport protein 4 (EC 6.2.1.-) (Fatty acid transport protein 4) (FATP-4) (Solute carrier family 27 member 4). | |||||
|
PTRF_HUMAN
|
||||||
| NC score | 0.007662 (rank : 47) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NZI2, O00535, Q6GMY1, Q96H74, Q9BT85, Q9HAP4 | Gene names | PTRF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polymerase I and transcript release factor (PTRF protein). | |||||
|
FHOD1_HUMAN
|
||||||
| NC score | 0.007370 (rank : 48) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y613, Q59F76, Q6Y1F2, Q76MS8, Q8N521 | Gene names | FHOD1, FHOS | |||
|
Domain Architecture |

|
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.003734 (rank : 49) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.002521 (rank : 50) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | -0.000547 (rank : 51) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||