Please be patient as the page loads
|
CCNT1_MOUSE
|
||||||
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CCNT1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992383 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60563, O60581 | Gene names | CCNT1 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT2_HUMAN
|
||||||
| θ value | 8.35901e-139 (rank : 3) | NC score | 0.958644 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
CCNK_HUMAN
|
||||||
| θ value | 2.96089e-35 (rank : 4) | NC score | 0.812881 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 5) | NC score | 0.702244 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CCNL2_MOUSE
|
||||||
| θ value | 1.42661e-21 (rank : 6) | NC score | 0.697169 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
CCNL2_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 7) | NC score | 0.689284 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
CCNL1_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 8) | NC score | 0.689930 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CCNL1_MOUSE
|
||||||
| θ value | 2.97466e-19 (rank : 9) | NC score | 0.691434 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
CCNC_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 10) | NC score | 0.619968 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P24863, Q9H543 | Gene names | CCNC | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-C. | |||||
|
CCNC_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 11) | NC score | 0.598808 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62447, Q9WUZ4 | Gene names | Ccnc | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-C. | |||||
|
CCNH_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 12) | NC score | 0.292716 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51946, Q8TBL9 | Gene names | CCNH | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-H (MO15-associated protein) (p37) (p34). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 13) | NC score | 0.096788 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.055933 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CCNA2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.094721 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNH_MOUSE
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.260489 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61458, Q9CVJ0, Q9JHV7 | Gene names | Ccnh | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-H. | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.063805 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.014789 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CD2L6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.014278 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BWU1, Q5JQZ7, Q5JR00, Q8TC78, Q9UPX2 | Gene names | CDC2L6, CDK11, KIAA1028 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 6 (EC 2.7.11.22) (CDC2- related protein kinase 6) (Death-preventing kinase) (Cyclin-dependent kinase 11). | |||||
|
DJBP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.052530 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
MK12_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.005868 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53778, Q14260, Q99588, Q99672 | Gene names | MAPK12, ERK6, SAPK3 | |||
|
Domain Architecture |
|
|||||
| Description | Mitogen-activated protein kinase 12 (EC 2.7.11.24) (Extracellular signal-regulated kinase 6) (ERK-6) (ERK5) (Stress-activated protein kinase 3) (Mitogen-activated protein kinase p38 gamma) (MAP kinase p38 gamma). | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.033820 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.006455 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MK12_MOUSE
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.005288 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08911, Q9D0M4 | Gene names | Mapk12, Sapk3 | |||
|
Domain Architecture |
|
|||||
| Description | Mitogen-activated protein kinase 12 (EC 2.7.11.24) (Extracellular signal-regulated kinase 6) (ERK-6) (Stress-activated protein kinase 3) (Mitogen-activated protein kinase p38 gamma) (MAP kinase p38 gamma). | |||||
|
UD110_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.015143 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HAW8, O00474, Q6NT91 | Gene names | UGT1A10, GNT1, UGT1 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucuronosyltransferase 1-10 precursor (EC 2.4.1.17) (UDPGT) (UGT1*10) (UGT1-10) (UGT1.10) (UGT-1J) (UGT1J). | |||||
|
AL2SA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.034637 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q53TS8, Q8NCN6, Q96LN4 | Gene names | ALS2CR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 11 protein. | |||||
|
CA026_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.025221 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T5J6, Q8NEK9, Q9BZQ7, Q9NXQ0 | Gene names | C1orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26. | |||||
|
CD2L6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.012003 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BWD8, Q80TM1 | Gene names | Cdc2l6, Kiaa1028 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 6 (EC 2.7.11.22) (CDC2- related protein kinase 6). | |||||
|
KIF17_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.008322 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
SAKS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.029522 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q922Y1, Q3UCP8 | Gene names | Saks1, D19Ertd721e | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein) (mY33K) (Protein 2B28). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.021488 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
UN13B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.012286 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.007037 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
COHA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.015303 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
DTNA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.012925 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
PAPPA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.030700 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R4K8, Q80VW3, Q80YY1, Q8K423, Q8R4K7, Q9ES06, Q9JK57 | Gene names | Pappa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
ZN438_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.008199 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
MYO5B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.003943 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.003385 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.047855 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
ING3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.012033 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NXR8, O60394, Q6GMT3, Q7Z762, Q969G0, Q96DT4, Q9HC99, Q9P081 | Gene names | ING3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
SRGP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.008702 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.008565 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
GLT12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.003808 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGT9 | Gene names | Galnt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.013236 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
PAPPA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.024618 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
PTPRV_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.006392 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.002726 (rank : 61) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.006911 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CABL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.022004 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3M5 | Gene names | Cables2 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
CB010_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.013968 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z570, Q6ZN26 | Gene names | C2orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf10. | |||||
|
CHCH3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.010699 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CRB9 | Gene names | Chchd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil-helix-coiled-coil-helix domain-containing protein 3. | |||||
|
NC6IP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.011378 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q923W1, Q6DI60, Q6PEA7, Q8R0W9 | Gene names | Ncoa6ip, Pimt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT). | |||||
|
PO4F3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.006101 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15319, O60557 | Gene names | POU4F3, BRN3C | |||
|
Domain Architecture |
|
|||||
| Description | POU domain, class 4, transcription factor 3 (Brain-specific homeobox/POU domain protein 3C) (Brn-3C). | |||||
|
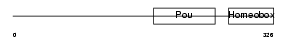
PO4F3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.006094 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63955 | Gene names | Pou4f3, Brn-3c, Brn3c | |||
|
Domain Architecture |
|
|||||
| Description | POU domain, class 4, transcription factor 3 (Brain-specific homeobox/POU domain protein 3C) (Brn-3C). | |||||
|
SAKS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.022187 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04323, Q9BV93, Q9BVV5 | Gene names | SAKS1 | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein). | |||||
|
SEM6D_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.003197 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.007464 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
CCNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.064600 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.059934 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |
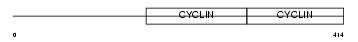
|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.055371 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNT1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT1_HUMAN
|
||||||
| NC score | 0.992383 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60563, O60581 | Gene names | CCNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT2_HUMAN
|
||||||
| NC score | 0.958644 (rank : 3) | θ value | 8.35901e-139 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
CCNK_HUMAN
|
||||||
| NC score | 0.812881 (rank : 4) | θ value | 2.96089e-35 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.702244 (rank : 5) | θ value | 1.12514e-34 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CCNL2_MOUSE
|
||||||
| NC score | 0.697169 (rank : 6) | θ value | 1.42661e-21 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
CCNL1_MOUSE
|
||||||
| NC score | 0.691434 (rank : 7) | θ value | 2.97466e-19 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
CCNL1_HUMAN
|
||||||
| NC score | 0.689930 (rank : 8) | θ value | 2.97466e-19 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CCNL2_HUMAN
|
||||||
| NC score | 0.689284 (rank : 9) | θ value | 1.5773e-20 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
CCNC_HUMAN
|
||||||
| NC score | 0.619968 (rank : 10) | θ value | 1.02475e-11 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P24863, Q9H543 | Gene names | CCNC | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-C. | |||||
|
CCNC_MOUSE
|
||||||
| NC score | 0.598808 (rank : 11) | θ value | 5.62301e-10 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62447, Q9WUZ4 | Gene names | Ccnc | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-C. | |||||
|
CCNH_HUMAN
|
||||||
| NC score | 0.292716 (rank : 12) | θ value | 0.0330416 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51946, Q8TBL9 | Gene names | CCNH | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-H (MO15-associated protein) (p37) (p34). | |||||
|
CCNH_MOUSE
|
||||||
| NC score | 0.260489 (rank : 13) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61458, Q9CVJ0, Q9JHV7 | Gene names | Ccnh | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-H. | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.096788 (rank : 14) | θ value | 0.0431538 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CCNA2_HUMAN
|
||||||
| NC score | 0.094721 (rank : 15) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA1_HUMAN
|
||||||
| NC score | 0.064600 (rank : 16) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.063805 (rank : 17) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNA1_MOUSE
|
||||||
| NC score | 0.059934 (rank : 18) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.055933 (rank : 19) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CCNA2_MOUSE
|
||||||
| NC score | 0.055371 (rank : 20) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
DJBP_HUMAN
|
||||||
| NC score | 0.052530 (rank : 21) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.047855 (rank : 22) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
AL2SA_HUMAN
|
||||||
| NC score | 0.034637 (rank : 23) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q53TS8, Q8NCN6, Q96LN4 | Gene names | ALS2CR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 11 protein. | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.033820 (rank : 24) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
PAPPA_MOUSE
|
||||||
| NC score | 0.030700 (rank : 25) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R4K8, Q80VW3, Q80YY1, Q8K423, Q8R4K7, Q9ES06, Q9JK57 | Gene names | Pappa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
SAKS1_MOUSE
|
||||||
| NC score | 0.029522 (rank : 26) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q922Y1, Q3UCP8 | Gene names | Saks1, D19Ertd721e | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein) (mY33K) (Protein 2B28). | |||||
|
CA026_HUMAN
|
||||||
| NC score | 0.025221 (rank : 27) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T5J6, Q8NEK9, Q9BZQ7, Q9NXQ0 | Gene names | C1orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26. | |||||
|
PAPPA_HUMAN
|
||||||
| NC score | 0.024618 (rank : 28) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
SAKS1_HUMAN
|
||||||
| NC score | 0.022187 (rank : 29) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04323, Q9BV93, Q9BVV5 | Gene names | SAKS1 | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein). | |||||
|
CABL2_MOUSE
|
||||||
| NC score | 0.022004 (rank : 30) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3M5 | Gene names | Cables2 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.021488 (rank : 31) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
COHA1_HUMAN
|
||||||
| NC score | 0.015303 (rank : 32) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
UD110_HUMAN
|
||||||
| NC score | 0.015143 (rank : 33) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HAW8, O00474, Q6NT91 | Gene names | UGT1A10, GNT1, UGT1 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucuronosyltransferase 1-10 precursor (EC 2.4.1.17) (UDPGT) (UGT1*10) (UGT1-10) (UGT1.10) (UGT-1J) (UGT1J). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.014789 (rank : 34) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CD2L6_HUMAN
|
||||||
| NC score | 0.014278 (rank : 35) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BWU1, Q5JQZ7, Q5JR00, Q8TC78, Q9UPX2 | Gene names | CDC2L6, CDK11, KIAA1028 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 6 (EC 2.7.11.22) (CDC2- related protein kinase 6) (Death-preventing kinase) (Cyclin-dependent kinase 11). | |||||
|
CB010_HUMAN
|
||||||
| NC score | 0.013968 (rank : 36) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z570, Q6ZN26 | Gene names | C2orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf10. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.013236 (rank : 37) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
DTNA_HUMAN
|
||||||
| NC score | 0.012925 (rank : 38) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
UN13B_HUMAN
|
||||||
| NC score | 0.012286 (rank : 39) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
ING3_HUMAN
|
||||||
| NC score | 0.012033 (rank : 40) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NXR8, O60394, Q6GMT3, Q7Z762, Q969G0, Q96DT4, Q9HC99, Q9P081 | Gene names | ING3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
CD2L6_MOUSE
|
||||||
| NC score | 0.012003 (rank : 41) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BWD8, Q80TM1 | Gene names | Cdc2l6, Kiaa1028 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 6 (EC 2.7.11.22) (CDC2- related protein kinase 6). | |||||
|
NC6IP_MOUSE
|
||||||
| NC score | 0.011378 (rank : 42) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q923W1, Q6DI60, Q6PEA7, Q8R0W9 | Gene names | Ncoa6ip, Pimt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT). | |||||
|
CHCH3_MOUSE
|
||||||
| NC score | 0.010699 (rank : 43) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CRB9 | Gene names | Chchd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil-helix-coiled-coil-helix domain-containing protein 3. | |||||
|
SRGP2_MOUSE
|
||||||
| NC score | 0.008702 (rank : 44) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.008565 (rank : 45) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
KIF17_MOUSE
|
||||||
| NC score | 0.008322 (rank : 46) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
ZN438_HUMAN
|
||||||
| NC score | 0.008199 (rank : 47) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.007464 (rank : 48) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.007037 (rank : 49) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.006911 (rank : 50) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.006455 (rank : 51) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
PTPRV_MOUSE
|
||||||
| NC score | 0.006392 (rank : 52) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
PO4F3_HUMAN
|
||||||
| NC score | 0.006101 (rank : 53) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15319, O60557 | Gene names | POU4F3, BRN3C | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 4, transcription factor 3 (Brain-specific homeobox/POU domain protein 3C) (Brn-3C). | |||||
|
PO4F3_MOUSE
|
||||||
| NC score | 0.006094 (rank : 54) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63955 | Gene names | Pou4f3, Brn-3c, Brn3c | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 4, transcription factor 3 (Brain-specific homeobox/POU domain protein 3C) (Brn-3C). | |||||
|
MK12_HUMAN
|
||||||
| NC score | 0.005868 (rank : 55) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53778, Q14260, Q99588, Q99672 | Gene names | MAPK12, ERK6, SAPK3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 12 (EC 2.7.11.24) (Extracellular signal-regulated kinase 6) (ERK-6) (ERK5) (Stress-activated protein kinase 3) (Mitogen-activated protein kinase p38 gamma) (MAP kinase p38 gamma). | |||||
|
MK12_MOUSE
|
||||||
| NC score | 0.005288 (rank : 56) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08911, Q9D0M4 | Gene names | Mapk12, Sapk3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 12 (EC 2.7.11.24) (Extracellular signal-regulated kinase 6) (ERK-6) (Stress-activated protein kinase 3) (Mitogen-activated protein kinase p38 gamma) (MAP kinase p38 gamma). | |||||
|
MYO5B_HUMAN
|
||||||
| NC score | 0.003943 (rank : 57) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
GLT12_MOUSE
|
||||||
| NC score | 0.003808 (rank : 58) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGT9 | Gene names | Galnt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.003385 (rank : 59) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
SEM6D_MOUSE
|
||||||
| NC score | 0.003197 (rank : 60) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.002726 (rank : 61) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||