Please be patient as the page loads
|
AL2SA_HUMAN
|
||||||
| SwissProt Accessions | Q53TS8, Q8NCN6, Q96LN4 | Gene names | ALS2CR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 11 protein. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AL2SA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q53TS8, Q8NCN6, Q96LN4 | Gene names | ALS2CR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 11 protein. | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 2) | NC score | 0.106077 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 3) | NC score | 0.054751 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
T3JAM_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 4) | NC score | 0.093262 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C0G2, Q8BY50, Q8K1Y9 | Gene names | Traf3ip3, T3jam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
APLP2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 5) | NC score | 0.077817 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06335 | Gene names | Aplp2 | |||
|
Domain Architecture |
|
|||||
| Description | Amyloid-like protein 2 precursor (CDEI box-binding protein) (CDEBP). | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 6) | NC score | 0.102983 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 7) | NC score | 0.031276 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.035674 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
BRCA1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.059021 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.057289 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.079470 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
TOPRS_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.069477 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.067898 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.076545 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
SPAG5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.066385 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.057234 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.082187 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
APBB1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.065398 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXJ1, O08642, Q3TPU0, Q8BNF4, Q8BSR9 | Gene names | Apbb1, Fe65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.050506 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.057146 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CLGN_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.042993 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14967 | Gene names | CLGN | |||
|
Domain Architecture |
|
|||||
| Description | Calmegin precursor. | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.072590 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
TAGAP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.041471 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
HDAC5_MOUSE
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.040532 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.036311 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.049224 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
APBB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.060481 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00213, Q96A93 | Gene names | APBB1, FE65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
JHD2C_MOUSE
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.042004 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
KAISO_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.010676 (rank : 103) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86T24, O00319, Q7Z361, Q8IVP6, Q8N3P0 | Gene names | ZBTB33, KAISO, ZNF348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional regulator Kaiso (Zinc finger and BTB domain-containing protein 33). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.062853 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.038071 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BICD2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.061645 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.062176 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.073734 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
KI21B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.042254 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.040266 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TAOK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.014679 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1172 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UL54, O94957, Q6UW73, Q7LC09, Q9NSW2 | Gene names | TAOK2, KIAA0881, MAP3K17, PSK, PSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2) (Prostate-derived STE20-like kinase 1) (PSK-1) (Kinase from chicken homolog C) (hKFC-C). | |||||
|
CCNT1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.034637 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
DBF4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.050682 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBU7, O75226, Q75MS6, Q75N01, Q9Y2M6 | Gene names | DBF4, ASK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DBF4 homolog (Activator of S phase Kinase). | |||||
|
DONS_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.048609 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXP4, Q80V93, Q8BV55 | Gene names | Donson, ORF60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein downstream neighbor of Son (Protein 3SG). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.045478 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
PTN14_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.025944 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
PTN14_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.025863 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
REST_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.044803 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
ZC11A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.048099 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
DHX29_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.019048 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
KI21B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.041017 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
LIPS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.083182 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54310, P97866, Q3TE34, Q6GU16 | Gene names | Lipe | |||
|
Domain Architecture |
|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.061199 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MRIP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.051343 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
TAOK2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.017632 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
TF2AY_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.044374 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UNN4 | Gene names | GTF2A1LF, ALF | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
UT14A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.042719 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BVJ6, Q5JYF1 | Gene names | UTP14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Antigen NY-CO- 16). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.013370 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
DBND2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.048245 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CRD4 | Gene names | Dbndd2, D2Bwg0891e | |||
|
Domain Architecture |
|
|||||
| Description | Dysbindin domain-containing protein 2. | |||||
|
FA29A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.051322 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z4H7, Q6IQ10, Q6NZX5, Q8IZQ4, Q96FN0, Q9H950, Q9H998, Q9HCJ8, Q9NXT8 | Gene names | FAM29A, KIAA1574 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM29A. | |||||
|
HOOK3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.046471 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.067706 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.053514 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PTN12_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.019854 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
RGP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.036470 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
SLK_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.016929 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SMRCD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.015373 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
|
CC025_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.046415 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NXP0, Q69YX4 | Gene names | C3orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf25. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.045696 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.054472 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DMXL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.022813 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.060111 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
RLF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.007286 (rank : 106) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13129, Q9NU60 | Gene names | RLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Rlf (Rearranged L-myc fusion gene protein) (Zn-15- related protein). | |||||
|
TFE2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.021673 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |
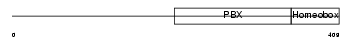
|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.048411 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.046379 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
ABCF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.027283 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.052170 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.026303 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
GBRA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.007550 (rank : 105) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48169 | Gene names | GABRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit alpha-4 precursor (GABA(A) receptor subunit alpha-4). | |||||
|
K1914_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.037523 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N4X5, Q8TB54, Q96PX4, Q96SY5 | Gene names | KIAA1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.042918 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
SIAS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.043427 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NR45, Q8WUV9, Q9BWS6, Q9NVD4 | Gene names | NANS, SAS | |||
|
Domain Architecture |
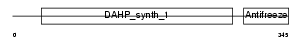
|
|||||
| Description | Sialic acid synthase (N-acetylneuraminate synthase) (EC 2.5.1.56) (N- acetylneuraminic acid synthase) (N-acetylneuraminate-9-phosphate synthase) (EC 2.5.1.57) (N-acetylneuraminic acid phosphate synthase). | |||||
|
SMCE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.033297 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
DCDC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.023034 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |
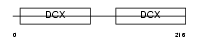
|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
ES8L2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.013907 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99K30, Q91VT7 | Gene names | Eps8l2, Eps8r2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
HOOK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.044569 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
KPB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.014579 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18826 | Gene names | Phka1 | |||
|
Domain Architecture |
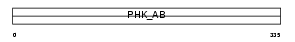
|
|||||
| Description | Phosphorylase b kinase regulatory subunit alpha, skeletal muscle isoform (Phosphorylase kinase alpha M subunit). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.032116 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
PDLI5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.014138 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.054750 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.037471 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
CN037_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.027609 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86TY3, Q6P5Q1, Q86TY1 | Gene names | C14orf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 precursor. | |||||
|
E41L5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.016165 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HCM4, Q9H975 | Gene names | EPB41L5, KIAA1548 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 5. | |||||
|
FGD6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.009830 (rank : 104) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
HDAC5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.031894 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.047281 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
PDLI5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.011012 (rank : 102) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CI51, Q9QYN0, Q9QYN1, Q9QYN2 | Gene names | Pdlim5, Enh | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
STIM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.024804 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9P246, Q96BF1, Q9BQH2, Q9H8R1 | Gene names | STIM2, KIAA1482 | |||
|
Domain Architecture |
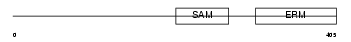
|
|||||
| Description | Stromal interaction molecule 2 precursor. | |||||
|
TOP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.021945 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q04750 | Gene names | Top1, Top-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.028790 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
AFF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.067515 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AFF3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.052040 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.072016 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.051981 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
BICD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.050678 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
BICD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.050587 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
MYST3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.052773 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.053935 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
T3JAM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.054975 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y228, Q2YDB5, Q4VY06, Q7Z706 | Gene names | TRAF3IP3, T3JAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
AL2SA_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q53TS8, Q8NCN6, Q96LN4 | Gene names | ALS2CR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 11 protein. | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.106077 (rank : 2) | θ value | 0.00390308 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.102983 (rank : 3) | θ value | 0.0961366 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
T3JAM_MOUSE
|
||||||
| NC score | 0.093262 (rank : 4) | θ value | 0.0252991 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C0G2, Q8BY50, Q8K1Y9 | Gene names | Traf3ip3, T3jam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
LIPS_MOUSE
|
||||||
| NC score | 0.083182 (rank : 5) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54310, P97866, Q3TE34, Q6GU16 | Gene names | Lipe | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.082187 (rank : 6) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.079470 (rank : 7) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
APLP2_MOUSE
|
||||||
| NC score | 0.077817 (rank : 8) | θ value | 0.0330416 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06335 | Gene names | Aplp2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (CDEI box-binding protein) (CDEBP). | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.076545 (rank : 9) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.073734 (rank : 10) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.072590 (rank : 11) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.072016 (rank : 12) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
TOPRS_HUMAN
|
||||||
| NC score | 0.069477 (rank : 13) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.067898 (rank : 14) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.067706 (rank : 15) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.067515 (rank : 16) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
SPAG5_HUMAN
|
||||||
| NC score | 0.066385 (rank : 17) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
APBB1_MOUSE
|
||||||
| NC score | 0.065398 (rank : 18) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXJ1, O08642, Q3TPU0, Q8BNF4, Q8BSR9 | Gene names | Apbb1, Fe65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.062853 (rank : 19) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.062176 (rank : 20) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
BICD2_HUMAN
|
||||||
| NC score | 0.061645 (rank : 21) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.061199 (rank : 22) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
APBB1_HUMAN
|
||||||
| NC score | 0.060481 (rank : 23) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00213, Q96A93 | Gene names | APBB1, FE65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.060111 (rank : 24) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
BRCA1_HUMAN
|
||||||
| NC score | 0.059021 (rank : 25) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.057289 (rank : 26) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.057234 (rank : 27) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.057146 (rank : 28) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
T3JAM_HUMAN
|
||||||
| NC score | 0.054975 (rank : 29) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y228, Q2YDB5, Q4VY06, Q7Z706 | Gene names | TRAF3IP3, T3JAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.054751 (rank : 30) | θ value | 0.0193708 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.054750 (rank : 31) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.054472 (rank : 32) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.053935 (rank : 33) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.053514 (rank : 34) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.052773 (rank : 35) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.052170 (rank : 36) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.052040 (rank : 37) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.051981 (rank : 38) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.051343 (rank : 39) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
FA29A_HUMAN
|
||||||
| NC score | 0.051322 (rank : 40) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z4H7, Q6IQ10, Q6NZX5, Q8IZQ4, Q96FN0, Q9H950, Q9H998, Q9HCJ8, Q9NXT8 | Gene names | FAM29A, KIAA1574 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM29A. | |||||
|
DBF4_HUMAN
|
||||||
| NC score | 0.050682 (rank : 41) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBU7, O75226, Q75MS6, Q75N01, Q9Y2M6 | Gene names | DBF4, ASK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DBF4 homolog (Activator of S phase Kinase). | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.050678 (rank : 42) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.050587 (rank : 43) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.050506 (rank : 44) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.049224 (rank : 45) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
DONS_MOUSE
|
||||||
| NC score | 0.048609 (rank : 46) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXP4, Q80V93, Q8BV55 | Gene names | Donson, ORF60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein downstream neighbor of Son (Protein 3SG). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.048411 (rank : 47) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
DBND2_MOUSE
|
||||||
| NC score | 0.048245 (rank : 48) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CRD4 | Gene names | Dbndd2, D2Bwg0891e | |||
|
Domain Architecture |

|
|||||
| Description | Dysbindin domain-containing protein 2. | |||||
|
ZC11A_MOUSE
|
||||||
| NC score | 0.048099 (rank : 49) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.047281 (rank : 50) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
HOOK3_MOUSE
|
||||||
| NC score | 0.046471 (rank : 51) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
CC025_HUMAN
|
||||||
| NC score | 0.046415 (rank : 52) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NXP0, Q69YX4 | Gene names | C3orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf25. | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.046379 (rank : 53) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.045696 (rank : 54) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.045478 (rank : 55) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.044803 (rank : 56) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
HOOK3_HUMAN
|
||||||
| NC score | 0.044569 (rank : 57) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
TF2AY_HUMAN
|
||||||
| NC score | 0.044374 (rank : 58) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UNN4 | Gene names | GTF2A1LF, ALF | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
SIAS_HUMAN
|
||||||
| NC score | 0.043427 (rank : 59) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NR45, Q8WUV9, Q9BWS6, Q9NVD4 | Gene names | NANS, SAS | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid synthase (N-acetylneuraminate synthase) (EC 2.5.1.56) (N- acetylneuraminic acid synthase) (N-acetylneuraminate-9-phosphate synthase) (EC 2.5.1.57) (N-acetylneuraminic acid phosphate synthase). | |||||
|
CLGN_HUMAN
|
||||||
| NC score | 0.042993 (rank : 60) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14967 | Gene names | CLGN | |||
|
Domain Architecture |

|
|||||
| Description | Calmegin precursor. | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.042918 (rank : 61) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
UT14A_HUMAN
|
||||||
| NC score | 0.042719 (rank : 62) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BVJ6, Q5JYF1 | Gene names | UTP14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Antigen NY-CO- 16). | |||||
|
KI21B_HUMAN
|
||||||
| NC score | 0.042254 (rank : 63) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
JHD2C_MOUSE
|
||||||
| NC score | 0.042004 (rank : 64) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
TAGAP_HUMAN
|
||||||
| NC score | 0.041471 (rank : 65) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
KI21B_MOUSE
|
||||||
| NC score | 0.041017 (rank : 66) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
HDAC5_MOUSE
|
||||||
| NC score | 0.040532 (rank : 67) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.040266 (rank : 68) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.038071 (rank : 69) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
K1914_HUMAN
|
||||||
| NC score | 0.037523 (rank : 70) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N4X5, Q8TB54, Q96PX4, Q96SY5 | Gene names | KIAA1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.037471 (rank : 71) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
RGP1_HUMAN
|
||||||
| NC score | 0.036470 (rank : 72) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.036311 (rank : 73) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.035674 (rank : 74) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CCNT1_MOUSE
|
||||||
| NC score | 0.034637 (rank : 75) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
SMCE1_HUMAN
|
||||||
| NC score | 0.033297 (rank : 76) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.032116 (rank : 77) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
HDAC5_HUMAN
|
||||||
| NC score | 0.031894 (rank : 78) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.031276 (rank : 79) | θ value | 0.125558 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.028790 (rank : 80) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
CN037_HUMAN
|
||||||
| NC score | 0.027609 (rank : 81) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86TY3, Q6P5Q1, Q86TY1 | Gene names | C14orf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 precursor. | |||||
|
ABCF1_HUMAN
|
||||||
| NC score | 0.027283 (rank : 82) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.026303 (rank : 83) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
PTN14_HUMAN
|
||||||
| NC score | 0.025944 (rank : 84) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
PTN14_MOUSE
|
||||||
| NC score | 0.025863 (rank : 85) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
STIM2_HUMAN
|
||||||
| NC score | 0.024804 (rank : 86) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9P246, Q96BF1, Q9BQH2, Q9H8R1 | Gene names | STIM2, KIAA1482 | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 2 precursor. | |||||
|
DCDC2_HUMAN
|
||||||
| NC score | 0.023034 (rank : 87) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |

|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
DMXL1_HUMAN
|
||||||
| NC score | 0.022813 (rank : 88) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
TOP1_MOUSE
|
||||||
| NC score | 0.021945 (rank : 89) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q04750 | Gene names | Top1, Top-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
TFE2_HUMAN
|
||||||
| NC score | 0.021673 (rank : 90) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
PTN12_MOUSE
|
||||||
| NC score | 0.019854 (rank : 91) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
DHX29_HUMAN
|
||||||
| NC score | 0.019048 (rank : 92) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
TAOK2_MOUSE
|
||||||
| NC score | 0.017632 (rank : 93) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.016929 (rank : 94) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
E41L5_HUMAN
|
||||||
| NC score | 0.016165 (rank : 95) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HCM4, Q9H975 | Gene names | EPB41L5, KIAA1548 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 5. | |||||
|
SMRCD_HUMAN
|
||||||
| NC score | 0.015373 (rank : 96) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
|
TAOK2_HUMAN
|
||||||
| NC score | 0.014679 (rank : 97) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1172 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UL54, O94957, Q6UW73, Q7LC09, Q9NSW2 | Gene names | TAOK2, KIAA0881, MAP3K17, PSK, PSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2) (Prostate-derived STE20-like kinase 1) (PSK-1) (Kinase from chicken homolog C) (hKFC-C). | |||||
|
KPB1_MOUSE
|
||||||
| NC score | 0.014579 (rank : 98) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18826 | Gene names | Phka1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylase b kinase regulatory subunit alpha, skeletal muscle isoform (Phosphorylase kinase alpha M subunit). | |||||
|
PDLI5_HUMAN
|
||||||
| NC score | 0.014138 (rank : 99) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
ES8L2_MOUSE
|
||||||
| NC score | 0.013907 (rank : 100) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99K30, Q91VT7 | Gene names | Eps8l2, Eps8r2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.013370 (rank : 101) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
PDLI5_MOUSE
|
||||||
| NC score | 0.011012 (rank : 102) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CI51, Q9QYN0, Q9QYN1, Q9QYN2 | Gene names | Pdlim5, Enh | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
KAISO_HUMAN
|
||||||
| NC score | 0.010676 (rank : 103) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86T24, O00319, Q7Z361, Q8IVP6, Q8N3P0 | Gene names | ZBTB33, KAISO, ZNF348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional regulator Kaiso (Zinc finger and BTB domain-containing protein 33). | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.009830 (rank : 104) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
GBRA4_HUMAN
|
||||||
| NC score | 0.007550 (rank : 105) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48169 | Gene names | GABRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit alpha-4 precursor (GABA(A) receptor subunit alpha-4). | |||||
|
RLF_HUMAN
|
||||||
| NC score | 0.007286 (rank : 106) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13129, Q9NU60 | Gene names | RLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Rlf (Rearranged L-myc fusion gene protein) (Zn-15- related protein). | |||||