Please be patient as the page loads
|
TF2AY_HUMAN
|
||||||
| SwissProt Accessions | Q9UNN4 | Gene names | GTF2A1LF, ALF | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TF2AY_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UNN4 | Gene names | GTF2A1LF, ALF | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
TF2AY_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.978057 (rank : 2) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R4I4, Q99PM2, Q9DAF4 | Gene names | Gtf2a1lf, Alf | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
TF2AA_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 3) | NC score | 0.788441 (rank : 4) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P52655 | Gene names | GTF2A1, TF2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) (TFIIA-42) (TFIIAL) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
TF2AA_MOUSE
|
||||||
| θ value | 1.52067e-31 (rank : 4) | NC score | 0.788959 (rank : 3) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PM3, Q8C812 | Gene names | Gtf2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 5) | NC score | 0.142406 (rank : 6) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 6) | NC score | 0.130150 (rank : 8) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 7) | NC score | 0.138498 (rank : 7) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.115163 (rank : 11) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
NEK1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.013337 (rank : 69) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1129 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96PY6, Q9Y594 | Gene names | NEK1, KIAA1901 | |||
|
Domain Architecture |
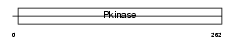
|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1) (NY-REN-55 antigen). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.095570 (rank : 13) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.048519 (rank : 36) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PCBP4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.047302 (rank : 37) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.062207 (rank : 25) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.056446 (rank : 27) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.123191 (rank : 9) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
PTMA_MOUSE
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.159893 (rank : 5) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.059232 (rank : 26) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
PCBP4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.043936 (rank : 41) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.064707 (rank : 24) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.119362 (rank : 10) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
CV106_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.056318 (rank : 28) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P0N0, Q86V14, Q96PY4, Q9NUR5, Q9Y4X9 | Gene names | C14orf106, KIAA1903 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf106 (P243). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.088211 (rank : 15) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.046773 (rank : 38) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.046558 (rank : 39) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.104250 (rank : 12) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.075401 (rank : 20) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
SUHW4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.016503 (rank : 62) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.050376 (rank : 33) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
AL2SA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.044374 (rank : 40) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q53TS8, Q8NCN6, Q96LN4 | Gene names | ALS2CR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 11 protein. | |||||
|
EAPP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.035939 (rank : 44) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q56P03, Q9BVF4, Q9NWV5, Q9NZ86 | Gene names | EAPP, C14orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E2F-associated phosphoprotein (EAPP). | |||||
|
ELF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.016104 (rank : 64) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.029995 (rank : 49) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.051064 (rank : 32) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
AN32B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.033525 (rank : 47) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.008649 (rank : 76) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.016240 (rank : 63) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.031872 (rank : 48) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
ABTAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.053713 (rank : 31) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.080445 (rank : 18) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
HNRPC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.022557 (rank : 56) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07910, P22628, Q53EX2, Q59FD3, Q5FWE8, Q86SF8, Q86U45, Q96HK7, Q96HM4, Q96IY5, Q9BTS3 | Gene names | HNRPC | |||
|
Domain Architecture |
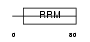
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.091694 (rank : 14) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.086118 (rank : 16) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.017267 (rank : 61) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
RBM28_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.025221 (rank : 53) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NW13, Q96CV3 | Gene names | RBM28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
TICN3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.014169 (rank : 68) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
UBP47_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.016026 (rank : 65) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
WDR43_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.023745 (rank : 54) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15061, Q15395, Q92577 | Gene names | WDR43, KIAA0007 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 43. | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.008361 (rank : 77) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.043452 (rank : 42) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
KCC2B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.003238 (rank : 82) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 841 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P28652, Q5SVH9 | Gene names | Camk2b, Camk2d | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II beta chain (EC 2.7.11.17) (CaM-kinase II beta chain) (CaM kinase II subunit beta) (CaMK-II subunit beta). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.027938 (rank : 51) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.018721 (rank : 58) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.023413 (rank : 55) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
PC11Y_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.003328 (rank : 81) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZA8, Q70LR6, Q70LR8, Q70LS0, Q70LS1, Q70LS2, Q70LS3, Q70LS4, Q70LS5, Q8WY34, Q9BZA9, Q9H4E1 | Gene names | PCDH11Y, PCDH11, PCDH22, PCDHY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 Y-linked precursor (Protocadherin-11) (Protocadherin- 22) (Protocadherin on the Y chromosome) (PCDH-Y) (Protocadherin prostate cancer) (Protocadherin-PC). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.039262 (rank : 43) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
RPA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.035365 (rank : 45) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
RPA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.028280 (rank : 50) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35134, Q7TSA9 | Gene names | Polr1a, Rpa1, Rpo1-4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.022058 (rank : 57) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CALX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.014967 (rank : 67) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
COCA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.004527 (rank : 79) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.070277 (rank : 21) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.034834 (rank : 46) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.027363 (rank : 52) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.017567 (rank : 60) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
ZN406_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.001060 (rank : 83) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TS63 | Gene names | Znf406, Zfp406 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406. | |||||
|
CFDP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.048662 (rank : 35) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UEE9, O00393, O00404, Q9UEF0, Q9UEF1, Q9UEF8 | Gene names | CFDP1, BCNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur). | |||||
|
GCP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.012921 (rank : 70) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96RT7, Q5JZ80, Q6PJ40, Q86YE9, Q9BY91, Q9UGX3, Q9UGX4 | Gene names | TUBGCP6, GCP6, KIAA1669 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 6 (GCP-6). | |||||
|
MN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.017757 (rank : 59) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
P3C2G_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.006581 (rank : 78) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PAX5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.009889 (rank : 73) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02548, O75933 | Gene names | PAX5 | |||
|
Domain Architecture |
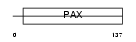
|
|||||
| Description | Paired box protein Pax-5 (B-cell-specific transcription factor) (BSAP). | |||||
|
PAX5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.009882 (rank : 74) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02650 | Gene names | Pax5, Pax-5 | |||
|
Domain Architecture |
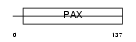
|
|||||
| Description | Paired box protein Pax-5 (B-cell-specific transcription factor) (BSAP). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.010018 (rank : 72) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
S6A13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.003591 (rank : 80) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NSD5 | Gene names | SLC6A13 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium- and chloride-dependent GABA transporter 2. | |||||
|
SEN34_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.015804 (rank : 66) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BSV6, Q9BVT1, Q9H6H5 | Gene names | TSEN34, LENG5, SEN34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (HsSen34) (Leukocyte receptor cluster member 5). | |||||
|
SNPC4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.012447 (rank : 71) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
SPAG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.008727 (rank : 75) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.049092 (rank : 34) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
CC47_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.066090 (rank : 22) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.076011 (rank : 19) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
MYT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.065078 (rank : 23) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
SG16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.083374 (rank : 17) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02815, Q62263 | Gene names | Spt1, Spt-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 16.5 kDa submandibular gland glycoprotein precursor (Salivary protein 1). | |||||
|
ST18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.056007 (rank : 30) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60284 | Gene names | ST18, KIAA0535, ZNF387 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18 (Zinc finger protein 387). | |||||
|
ST18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.056052 (rank : 29) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TY4, Q3UH00, Q3URH9, Q3UVB9, Q3UZN9, Q811B4, Q8K098 | Gene names | St18, Kiaa0535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18. | |||||
|
TF2AY_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UNN4 | Gene names | GTF2A1LF, ALF | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
TF2AY_MOUSE
|
||||||
| NC score | 0.978057 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R4I4, Q99PM2, Q9DAF4 | Gene names | Gtf2a1lf, Alf | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
TF2AA_MOUSE
|
||||||
| NC score | 0.788959 (rank : 3) | θ value | 1.52067e-31 (rank : 4) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PM3, Q8C812 | Gene names | Gtf2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
TF2AA_HUMAN
|
||||||
| NC score | 0.788441 (rank : 4) | θ value | 1.52067e-31 (rank : 3) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P52655 | Gene names | GTF2A1, TF2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) (TFIIA-42) (TFIIAL) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.159893 (rank : 5) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.142406 (rank : 6) | θ value | 0.000602161 (rank : 5) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.138498 (rank : 7) | θ value | 0.00509761 (rank : 7) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.130150 (rank : 8) | θ value | 0.000602161 (rank : 6) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.123191 (rank : 9) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.119362 (rank : 10) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.115163 (rank : 11) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.104250 (rank : 12) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.095570 (rank : 13) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.091694 (rank : 14) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.088211 (rank : 15) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.086118 (rank : 16) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
SG16_MOUSE
|
||||||
| NC score | 0.083374 (rank : 17) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02815, Q62263 | Gene names | Spt1, Spt-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 16.5 kDa submandibular gland glycoprotein precursor (Salivary protein 1). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.080445 (rank : 18) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.076011 (rank : 19) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.075401 (rank : 20) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.070277 (rank : 21) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.066090 (rank : 22) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.065078 (rank : 23) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.064707 (rank : 24) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.062207 (rank : 25) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.059232 (rank : 26) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.056446 (rank : 27) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
CV106_HUMAN
|
||||||
| NC score | 0.056318 (rank : 28) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P0N0, Q86V14, Q96PY4, Q9NUR5, Q9Y4X9 | Gene names | C14orf106, KIAA1903 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf106 (P243). | |||||
|
ST18_MOUSE
|
||||||
| NC score | 0.056052 (rank : 29) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TY4, Q3UH00, Q3URH9, Q3UVB9, Q3UZN9, Q811B4, Q8K098 | Gene names | St18, Kiaa0535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18. | |||||
|
ST18_HUMAN
|
||||||
| NC score | 0.056007 (rank : 30) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60284 | Gene names | ST18, KIAA0535, ZNF387 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18 (Zinc finger protein 387). | |||||
|
ABTAP_HUMAN
|
||||||
| NC score | 0.053713 (rank : 31) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.051064 (rank : 32) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.050376 (rank : 33) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.049092 (rank : 34) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
CFDP1_HUMAN
|
||||||
| NC score | 0.048662 (rank : 35) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UEE9, O00393, O00404, Q9UEF0, Q9UEF1, Q9UEF8 | Gene names | CFDP1, BCNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.048519 (rank : 36) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PCBP4_MOUSE
|
||||||
| NC score | 0.047302 (rank : 37) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.046773 (rank : 38) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.046558 (rank : 39) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
AL2SA_HUMAN
|
||||||
| NC score | 0.044374 (rank : 40) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q53TS8, Q8NCN6, Q96LN4 | Gene names | ALS2CR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 11 protein. | |||||
|
PCBP4_HUMAN
|
||||||
| NC score | 0.043936 (rank : 41) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.043452 (rank : 42) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.039262 (rank : 43) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
EAPP_HUMAN
|
||||||
| NC score | 0.035939 (rank : 44) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q56P03, Q9BVF4, Q9NWV5, Q9NZ86 | Gene names | EAPP, C14orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E2F-associated phosphoprotein (EAPP). | |||||
|
RPA1_HUMAN
|
||||||
| NC score | 0.035365 (rank : 45) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.034834 (rank : 46) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
AN32B_MOUSE
|
||||||
| NC score | 0.033525 (rank : 47) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.031872 (rank : 48) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.029995 (rank : 49) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RPA1_MOUSE
|
||||||
| NC score | 0.028280 (rank : 50) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35134, Q7TSA9 | Gene names | Polr1a, Rpa1, Rpo1-4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.027938 (rank : 51) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.027363 (rank : 52) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
RBM28_HUMAN
|
||||||
| NC score | 0.025221 (rank : 53) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NW13, Q96CV3 | Gene names | RBM28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
WDR43_HUMAN
|
||||||
| NC score | 0.023745 (rank : 54) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15061, Q15395, Q92577 | Gene names | WDR43, KIAA0007 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 43. | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.023413 (rank : 55) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
HNRPC_HUMAN
|
||||||
| NC score | 0.022557 (rank : 56) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07910, P22628, Q53EX2, Q59FD3, Q5FWE8, Q86SF8, Q86U45, Q96HK7, Q96HM4, Q96IY5, Q9BTS3 | Gene names | HNRPC | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.022058 (rank : 57) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.018721 (rank : 58) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
MN1_HUMAN
|
||||||
| NC score | 0.017757 (rank : 59) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.017567 (rank : 60) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.017267 (rank : 61) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
SUHW4_HUMAN
|
||||||
| NC score | 0.016503 (rank : 62) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.016240 (rank : 63) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
ELF4_HUMAN
|
||||||
| NC score | 0.016104 (rank : 64) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
UBP47_MOUSE
|
||||||
| NC score | 0.016026 (rank : 65) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
SEN34_HUMAN
|
||||||
| NC score | 0.015804 (rank : 66) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BSV6, Q9BVT1, Q9H6H5 | Gene names | TSEN34, LENG5, SEN34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (HsSen34) (Leukocyte receptor cluster member 5). | |||||
|
CALX_MOUSE
|
||||||
| NC score | 0.014967 (rank : 67) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
TICN3_MOUSE
|
||||||
| NC score | 0.014169 (rank : 68) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
NEK1_HUMAN
|
||||||
| NC score | 0.013337 (rank : 69) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1129 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96PY6, Q9Y594 | Gene names | NEK1, KIAA1901 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1) (NY-REN-55 antigen). | |||||
|
GCP6_HUMAN
|
||||||
| NC score | 0.012921 (rank : 70) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96RT7, Q5JZ80, Q6PJ40, Q86YE9, Q9BY91, Q9UGX3, Q9UGX4 | Gene names | TUBGCP6, GCP6, KIAA1669 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 6 (GCP-6). | |||||
|
SNPC4_MOUSE
|
||||||
| NC score | 0.012447 (rank : 71) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.010018 (rank : 72) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
PAX5_HUMAN
|
||||||
| NC score | 0.009889 (rank : 73) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02548, O75933 | Gene names | PAX5 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-5 (B-cell-specific transcription factor) (BSAP). | |||||
|
PAX5_MOUSE
|
||||||
| NC score | 0.009882 (rank : 74) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02650 | Gene names | Pax5, Pax-5 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-5 (B-cell-specific transcription factor) (BSAP). | |||||
|
SPAG1_MOUSE
|
||||||
| NC score | 0.008727 (rank : 75) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.008649 (rank : 76) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.008361 (rank : 77) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
P3C2G_MOUSE
|
||||||
| NC score | 0.006581 (rank : 78) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
COCA1_HUMAN
|
||||||
| NC score | 0.004527 (rank : 79) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
S6A13_HUMAN
|
||||||
| NC score | 0.003591 (rank : 80) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NSD5 | Gene names | SLC6A13 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium- and chloride-dependent GABA transporter 2. | |||||
|
PC11Y_HUMAN
|
||||||
| NC score | 0.003328 (rank : 81) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZA8, Q70LR6, Q70LR8, Q70LS0, Q70LS1, Q70LS2, Q70LS3, Q70LS4, Q70LS5, Q8WY34, Q9BZA9, Q9H4E1 | Gene names | PCDH11Y, PCDH11, PCDH22, PCDHY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 Y-linked precursor (Protocadherin-11) (Protocadherin- 22) (Protocadherin on the Y chromosome) (PCDH-Y) (Protocadherin prostate cancer) (Protocadherin-PC). | |||||
|
KCC2B_MOUSE
|
||||||
| NC score | 0.003238 (rank : 82) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 841 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P28652, Q5SVH9 | Gene names | Camk2b, Camk2d | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II beta chain (EC 2.7.11.17) (CaM-kinase II beta chain) (CaM kinase II subunit beta) (CaMK-II subunit beta). | |||||
|
ZN406_MOUSE
|
||||||
| NC score | 0.001060 (rank : 83) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TS63 | Gene names | Znf406, Zfp406 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406. | |||||