Please be patient as the page loads
|
CFDP1_HUMAN
|
||||||
| SwissProt Accessions | Q9UEE9, O00393, O00404, Q9UEF0, Q9UEF1, Q9UEF8 | Gene names | CFDP1, BCNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CFDP1_HUMAN
|
||||||
| θ value | 9.58617e-127 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9UEE9, O00393, O00404, Q9UEF0, Q9UEF1, Q9UEF8 | Gene names | CFDP1, BCNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur). | |||||
|
CFDP1_MOUSE
|
||||||
| θ value | 1.59114e-105 (rank : 2) | NC score | 0.946421 (rank : 2) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88271, O70565, Q9JMA5 | Gene names | Cfdp1, Bcnt, Cfdp, Cp27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur) (27 kDa craniofacial protein) (Protein Cp27). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 3) | NC score | 0.141394 (rank : 3) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
SEMG1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 4) | NC score | 0.106849 (rank : 7) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04279, Q6X4I9, Q6Y809, Q6Y822, Q6Y823, Q86U64, Q96QM3 | Gene names | SEMG1, SEMG | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-1 precursor (Semenogelin I) (SGI) [Contains: Alpha- inhibin-92; Alpha-inhibin-31; Seminal basic protein]. | |||||
|
K1210_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 5) | NC score | 0.134032 (rank : 4) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
APC_HUMAN
|
||||||
| θ value | 0.163984 (rank : 6) | NC score | 0.068924 (rank : 26) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 7) | NC score | 0.070667 (rank : 23) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.066600 (rank : 29) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 9) | NC score | 0.119420 (rank : 5) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.113229 (rank : 6) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
ERCC5_MOUSE
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.064369 (rank : 32) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.076520 (rank : 18) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.062891 (rank : 35) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
FEZ1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.090596 (rank : 9) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99689, O00679, O00728, Q6IBI7 | Gene names | FEZ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 1 (Zygin-1) (Zygin I). | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.068657 (rank : 27) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.088948 (rank : 12) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
FEZ1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.099162 (rank : 8) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K0X8 | Gene names | Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 1 (Zygin-1) (Zygin I). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.021726 (rank : 94) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.078075 (rank : 16) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.045141 (rank : 59) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.053289 (rank : 43) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.054768 (rank : 39) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
NEB2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.043400 (rank : 62) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.038976 (rank : 68) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TF2AY_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.064137 (rank : 33) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R4I4, Q99PM2, Q9DAF4 | Gene names | Gtf2a1lf, Alf | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
TIM44_MOUSE
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.053033 (rank : 44) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35857 | Gene names | Timm44, Mimt44, Tim44 | |||
|
Domain Architecture |

|
|||||
| Description | Import inner membrane translocase subunit TIM44, mitochondrial precursor. | |||||
|
ARD1H_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.048277 (rank : 55) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41227 | Gene names | ARD1A, ARD1, TE2 | |||
|
Domain Architecture |
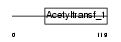
|
|||||
| Description | N-terminal acetyltransferase complex ARD1 subunit homolog A (EC 2.3.1.88) (EC 2.3.1.-). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.052526 (rank : 46) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
GP179_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.045032 (rank : 60) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.023604 (rank : 88) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
ZHX2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.030101 (rank : 82) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.030313 (rank : 81) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.048623 (rank : 54) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.034938 (rank : 73) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.040303 (rank : 65) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.067415 (rank : 28) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.077575 (rank : 17) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.033532 (rank : 77) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.082758 (rank : 14) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.038789 (rank : 69) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
PA24A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.022182 (rank : 92) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47713 | Gene names | Pla2g4a, Cpla2, Pla2g4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.037800 (rank : 72) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
LIPB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.043685 (rank : 61) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86W92, O75336, Q86X70, Q9NY03, Q9ULJ0 | Gene names | PPFIBP1, KIAA1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1) (hSGT2). | |||||
|
LIPB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.039276 (rank : 67) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
NEB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.038561 (rank : 70) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.047097 (rank : 57) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TULP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.033618 (rank : 76) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z273 | Gene names | Tulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
APXL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.033400 (rank : 78) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.040871 (rank : 64) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.034760 (rank : 74) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
GLT14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.008969 (rank : 99) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BVG5, Q60GT2, Q8BTI6, Q9DCJ2 | Gene names | Galnt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
GP107_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.023178 (rank : 89) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VW38, Q5JPA3, Q5VW39, Q96T26, Q9H658, Q9HCE8 | Gene names | GPR107, KIAA1624, LUSTR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein GPR107 precursor (Lung seven transmembrane receptor 1). | |||||
|
RADI_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.028102 (rank : 85) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.034403 (rank : 75) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
EZH1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.021982 (rank : 93) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
GRP75_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.016675 (rank : 97) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
GRP75_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.015348 (rank : 98) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.030897 (rank : 80) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
NP1L3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.051935 (rank : 48) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99457, O60788 | Gene names | NAP1L3, BNAP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 3. | |||||
|
RBP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.028665 (rank : 84) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RBP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.027717 (rank : 86) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.039905 (rank : 66) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.070522 (rank : 24) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.090592 (rank : 10) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MIA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.024397 (rank : 87) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96PC5, Q9H6C1 | Gene names | MIA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.042511 (rank : 63) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.047676 (rank : 56) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TLK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.007874 (rank : 100) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C0V0 | Gene names | Tlk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1). | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.019095 (rank : 95) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
CD2L5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.003141 (rank : 102) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14004, Q6DKQ9, Q75MH4, Q75MH5, Q96JN4, Q9H4A0, Q9H4A1, Q9UDR4 | Gene names | CDC2L5, CDC2L, CHED, KIAA1791 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5) (Cholinesterase-related cell division controller). | |||||
|
DBF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.018932 (rank : 96) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBU7, O75226, Q75MS6, Q75N01, Q9Y2M6 | Gene names | DBF4, ASK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DBF4 homolog (Activator of S phase Kinase). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.022850 (rank : 90) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.022433 (rank : 91) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYRIP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.029239 (rank : 83) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
SCG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.047017 (rank : 58) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P13521, Q8TBH3 | Gene names | SCG2, CHGC | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.038156 (rank : 71) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.031516 (rank : 79) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TF2AY_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.048662 (rank : 53) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UNN4 | Gene names | GTF2A1LF, ALF | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
TLK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.007339 (rank : 101) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKI8, Q14150, Q8N591, Q9NYH2, Q9Y4F6 | Gene names | TLK1, KIAA0137 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1) (PKU-beta). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.050728 (rank : 51) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
APC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.055240 (rank : 38) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.052012 (rank : 47) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.050499 (rank : 52) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.064069 (rank : 34) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.074877 (rank : 19) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.065249 (rank : 31) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FA21C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.055803 (rank : 36) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.053786 (rank : 41) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
IF2P_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.052529 (rank : 45) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.071675 (rank : 22) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.072727 (rank : 20) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.051819 (rank : 49) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.066544 (rank : 30) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.070077 (rank : 25) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.088757 (rank : 13) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.089248 (rank : 11) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PTMS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.071704 (rank : 21) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
PTMS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.082230 (rank : 15) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
SEMG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.053606 (rank : 42) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.051798 (rank : 50) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.055458 (rank : 37) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.054305 (rank : 40) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
CFDP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 9.58617e-127 (rank : 1) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9UEE9, O00393, O00404, Q9UEF0, Q9UEF1, Q9UEF8 | Gene names | CFDP1, BCNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur). | |||||
|
CFDP1_MOUSE
|
||||||
| NC score | 0.946421 (rank : 2) | θ value | 1.59114e-105 (rank : 2) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88271, O70565, Q9JMA5 | Gene names | Cfdp1, Bcnt, Cfdp, Cp27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur) (27 kDa craniofacial protein) (Protein Cp27). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.141394 (rank : 3) | θ value | 0.00102713 (rank : 3) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
K1210_HUMAN
|
||||||
| NC score | 0.134032 (rank : 4) | θ value | 0.0961366 (rank : 5) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.119420 (rank : 5) | θ value | 0.21417 (rank : 9) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.113229 (rank : 6) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
SEMG1_HUMAN
|
||||||
| NC score | 0.106849 (rank : 7) | θ value | 0.0563607 (rank : 4) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04279, Q6X4I9, Q6Y809, Q6Y822, Q6Y823, Q86U64, Q96QM3 | Gene names | SEMG1, SEMG | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-1 precursor (Semenogelin I) (SGI) [Contains: Alpha- inhibin-92; Alpha-inhibin-31; Seminal basic protein]. | |||||
|
FEZ1_MOUSE
|
||||||
| NC score | 0.099162 (rank : 8) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K0X8 | Gene names | Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 1 (Zygin-1) (Zygin I). | |||||
|
FEZ1_HUMAN
|
||||||
| NC score | 0.090596 (rank : 9) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99689, O00679, O00728, Q6IBI7 | Gene names | FEZ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 1 (Zygin-1) (Zygin I). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.090592 (rank : 10) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.089248 (rank : 11) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.088948 (rank : 12) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.088757 (rank : 13) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.082758 (rank : 14) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.082230 (rank : 15) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.078075 (rank : 16) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.077575 (rank : 17) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.076520 (rank : 18) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.074877 (rank : 19) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.072727 (rank : 20) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.071704 (rank : 21) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.071675 (rank : 22) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.070667 (rank : 23) | θ value | 0.163984 (rank : 7) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.070522 (rank : 24) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.070077 (rank : 25) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.068924 (rank : 26) | θ value | 0.163984 (rank : 6) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.068657 (rank : 27) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.067415 (rank : 28) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.066600 (rank : 29) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.066544 (rank : 30) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.065249 (rank : 31) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ERCC5_MOUSE
|
||||||
| NC score | 0.064369 (rank : 32) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
TF2AY_MOUSE
|
||||||
| NC score | 0.064137 (rank : 33) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R4I4, Q99PM2, Q9DAF4 | Gene names | Gtf2a1lf, Alf | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.064069 (rank : 34) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.062891 (rank : 35) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.055803 (rank : 36) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.055458 (rank : 37) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.055240 (rank : 38) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.054768 (rank : 39) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.054305 (rank : 40) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.053786 (rank : 41) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
SEMG2_HUMAN
|
||||||
| NC score | 0.053606 (rank : 42) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.053289 (rank : 43) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
TIM44_MOUSE
|
||||||
| NC score | 0.053033 (rank : 44) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35857 | Gene names | Timm44, Mimt44, Tim44 | |||
|
Domain Architecture |

|
|||||
| Description | Import inner membrane translocase subunit TIM44, mitochondrial precursor. | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.052529 (rank : 45) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.052526 (rank : 46) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.052012 (rank : 47) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NP1L3_HUMAN
|
||||||
| NC score | 0.051935 (rank : 48) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99457, O60788 | Gene names | NAP1L3, BNAP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 3. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.051819 (rank : 49) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.051798 (rank : 50) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.050728 (rank : 51) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.050499 (rank : 52) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
TF2AY_HUMAN
|
||||||
| NC score | 0.048662 (rank : 53) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UNN4 | Gene names | GTF2A1LF, ALF | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.048623 (rank : 54) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
ARD1H_HUMAN
|
||||||
| NC score | 0.048277 (rank : 55) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41227 | Gene names | ARD1A, ARD1, TE2 | |||
|
Domain Architecture |

|
|||||
| Description | N-terminal acetyltransferase complex ARD1 subunit homolog A (EC 2.3.1.88) (EC 2.3.1.-). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.047676 (rank : 56) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.047097 (rank : 57) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
SCG2_HUMAN
|
||||||
| NC score | 0.047017 (rank : 58) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P13521, Q8TBH3 | Gene names | SCG2, CHGC | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.045141 (rank : 59) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.045032 (rank : 60) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
LIPB1_HUMAN
|
||||||
| NC score | 0.043685 (rank : 61) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86W92, O75336, Q86X70, Q9NY03, Q9ULJ0 | Gene names | PPFIBP1, KIAA1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1) (hSGT2). | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.043400 (rank : 62) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.042511 (rank : 63) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.040871 (rank : 64) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.040303 (rank : 65) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.039905 (rank : 66) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
LIPB1_MOUSE
|
||||||
| NC score | 0.039276 (rank : 67) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.038976 (rank : 68) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.038789 (rank : 69) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.038561 (rank : 70) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.038156 (rank : 71) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.037800 (rank : 72) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.034938 (rank : 73) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.034760 (rank : 74) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.034403 (rank : 75) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
TULP1_MOUSE
|
||||||
| NC score | 0.033618 (rank : 76) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z273 | Gene names | Tulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.033532 (rank : 77) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.033400 (rank : 78) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.031516 (rank : 79) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.030897 (rank : 80) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.030313 (rank : 81) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
ZHX2_MOUSE
|
||||||
| NC score | 0.030101 (rank : 82) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
MYRIP_MOUSE
|
||||||
| NC score | 0.029239 (rank : 83) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
RBP1_HUMAN
|
||||||
| NC score | 0.028665 (rank : 84) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.028102 (rank : 85) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.027717 (rank : 86) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
MIA2_HUMAN
|
||||||
| NC score | 0.024397 (rank : 87) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96PC5, Q9H6C1 | Gene names | MIA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.023604 (rank : 88) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
GP107_HUMAN
|
||||||
| NC score | 0.023178 (rank : 89) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VW38, Q5JPA3, Q5VW39, Q96T26, Q9H658, Q9HCE8 | Gene names | GPR107, KIAA1624, LUSTR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein GPR107 precursor (Lung seven transmembrane receptor 1). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.022850 (rank : 90) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.022433 (rank : 91) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
PA24A_MOUSE
|
||||||
| NC score | 0.022182 (rank : 92) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47713 | Gene names | Pla2g4a, Cpla2, Pla2g4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
EZH1_HUMAN
|
||||||
| NC score | 0.021982 (rank : 93) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.021726 (rank : 94) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.019095 (rank : 95) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
DBF4_HUMAN
|
||||||
| NC score | 0.018932 (rank : 96) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBU7, O75226, Q75MS6, Q75N01, Q9Y2M6 | Gene names | DBF4, ASK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DBF4 homolog (Activator of S phase Kinase). | |||||
|
GRP75_HUMAN
|
||||||
| NC score | 0.016675 (rank : 97) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
GRP75_MOUSE
|
||||||
| NC score | 0.015348 (rank : 98) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
GLT14_MOUSE
|
||||||
| NC score | 0.008969 (rank : 99) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BVG5, Q60GT2, Q8BTI6, Q9DCJ2 | Gene names | Galnt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
TLK1_MOUSE
|
||||||
| NC score | 0.007874 (rank : 100) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C0V0 | Gene names | Tlk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1). | |||||
|
TLK1_HUMAN
|
||||||
| NC score | 0.007339 (rank : 101) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKI8, Q14150, Q8N591, Q9NYH2, Q9Y4F6 | Gene names | TLK1, KIAA0137 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1) (PKU-beta). | |||||
|
CD2L5_HUMAN
|
||||||
| NC score | 0.003141 (rank : 102) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14004, Q6DKQ9, Q75MH4, Q75MH5, Q96JN4, Q9H4A0, Q9H4A1, Q9UDR4 | Gene names | CDC2L5, CDC2L, CHED, KIAA1791 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5) (Cholinesterase-related cell division controller). | |||||