Please be patient as the page loads
|
EZH1_HUMAN
|
||||||
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EZH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998109 (rank : 2) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | P70351, Q9R089 | Gene names | Ezh1, Enx2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.988594 (rank : 3) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q15910, Q15755, Q92857 | Gene names | EZH2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
EZH2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.988237 (rank : 4) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q61188, Q9R090 | Gene names | Ezh2, Enx1h | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
SUV91_HUMAN
|
||||||
| θ value | 2.43343e-21 (rank : 5) | NC score | 0.702841 (rank : 5) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
SUV91_MOUSE
|
||||||
| θ value | 2.43343e-21 (rank : 6) | NC score | 0.696594 (rank : 6) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 3.17815e-21 (rank : 7) | NC score | 0.555914 (rank : 15) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 4.58923e-20 (rank : 8) | NC score | 0.612885 (rank : 10) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 5.99374e-20 (rank : 9) | NC score | 0.564428 (rank : 13) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 10) | NC score | 0.651921 (rank : 9) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SUV92_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 11) | NC score | 0.691193 (rank : 7) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 1.33526e-19 (rank : 12) | NC score | 0.561693 (rank : 14) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 13) | NC score | 0.415198 (rank : 22) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 14) | NC score | 0.437011 (rank : 20) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
SUV92_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 15) | NC score | 0.684997 (rank : 8) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 16) | NC score | 0.537792 (rank : 16) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 17) | NC score | 0.422613 (rank : 21) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 5.60996e-18 (rank : 18) | NC score | 0.341415 (rank : 24) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 19) | NC score | 0.343227 (rank : 23) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EHMT1_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 20) | NC score | 0.334046 (rank : 25) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
SETD8_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 21) | NC score | 0.611168 (rank : 11) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
SETD8_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 22) | NC score | 0.610503 (rank : 12) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q2YDW7, Q8C0J9 | Gene names | SETD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8). | |||||
|
SETB1_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 23) | NC score | 0.474638 (rank : 18) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SETB1_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 24) | NC score | 0.469558 (rank : 19) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15047, Q96GM9 | Gene names | SETDB1, KIAA0067 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SETB2_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 25) | NC score | 0.509735 (rank : 17) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
SETD7_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 26) | NC score | 0.207438 (rank : 26) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VHL1 | Gene names | Setd7, Set7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7). | |||||
|
SETD7_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 27) | NC score | 0.201830 (rank : 27) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WTS6, Q9C0E6 | Gene names | SETD7, KIAA1717, SET7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7) (Set9) (SET7/9). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.133721 (rank : 29) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
ITB2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.036853 (rank : 106) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P11835, Q64482 | Gene names | Itgb2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/P150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.084501 (rank : 36) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.100161 (rank : 31) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
MCSP_MOUSE
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.158880 (rank : 28) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
PZRN3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.026737 (rank : 123) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
KRA44_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.065811 (rank : 54) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.042842 (rank : 96) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
KRA45_HUMAN
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.061230 (rank : 61) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
KIF4A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.017720 (rank : 134) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.046268 (rank : 93) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
TNR7_MOUSE
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.046799 (rank : 91) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P41272 | Gene names | Tnfrsf7, Cd27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.045559 (rank : 94) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
KRA24_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.082295 (rank : 40) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
NP1L1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.030665 (rank : 114) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28656, Q3UL14 | Gene names | Nap1l1, Nrp | |||
|
Domain Architecture |
|
|||||
| Description | Nucleosome assembly protein 1-like 1 (NAP-1-related protein) (Brain protein DN38). | |||||
|
PTMA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.097708 (rank : 32) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
CCD16_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.039458 (rank : 101) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96NB3, Q96F60, Q96GZ5, Q9BU38 | Gene names | CCDC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16. | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.053900 (rank : 79) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.038877 (rank : 102) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
PZRN3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.022621 (rank : 128) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.000313 (rank : 160) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
SEPP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.035366 (rank : 108) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49908, Q6PD59, Q6PI43, Q6PI87, Q6PJF9 | Gene names | SEPP1, SELP | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein P precursor (SeP). | |||||
|
STAB2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.025837 (rank : 124) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
TRIO_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.001533 (rank : 159) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
KR102_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.048079 (rank : 89) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KRA93_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.047527 (rank : 90) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
SMYD3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.070181 (rank : 48) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CWR2, Q6P7V6, Q8BG90 | Gene names | Smyd3, Zmynd1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.039512 (rank : 100) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.041878 (rank : 98) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
ITB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.038706 (rank : 103) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P05556, P78466, P78467, Q13089, Q13090, Q13091, Q13212, Q14622, Q14647 | Gene names | ITGB1, FNRB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
ZNFX1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.052236 (rank : 82) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
ITB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.030174 (rank : 116) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P05107, Q16418 | Gene names | ITGB2, CD18 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/p150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
KR511_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.094522 (rank : 33) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.066334 (rank : 51) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KRA94_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.052886 (rank : 80) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BYQ2 | Gene names | KRTAP9-4, KAP9.4, KRTAP9.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-4 (Keratin-associated protein 9.4) (Ultrahigh sulfur keratin-associated protein 9.4). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.046678 (rank : 92) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NOTC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.034805 (rank : 110) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NRDC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.015735 (rank : 138) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
SREC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.031960 (rank : 112) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
TAF5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.007854 (rank : 148) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15542, Q86UZ7, Q9Y4K5 | Gene names | TAF5, TAF2D | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 5 (Transcription initiation factor TFIID 100 kDa subunit) (TAF(II)100) (TAFII-100) (TAFII100). | |||||
|
TAF5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.006624 (rank : 149) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C092, Q8BTR2 | Gene names | Taf5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 5 (Transcription initiation factor TFIID 100 kDa subunit) (TAF(II)100) (TAFII-100) (TAFII100). | |||||
|
VWF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.035191 (rank : 109) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
ACY3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.021242 (rank : 131) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96HD9 | Gene names | ACY3, ASPA2 | |||
|
Domain Architecture |
|
|||||
| Description | Aspartoacylase-2 (EC 3.5.1.15) (Aminoacylase-3) (ACY-3) (Acylase III) (Hepatitis C virus core-binding protein 1) (HCBP1). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.016191 (rank : 137) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
KR101_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.038353 (rank : 104) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR103_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.050630 (rank : 84) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
KR105_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.055841 (rank : 72) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.012783 (rank : 140) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
SMYD3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.070677 (rank : 46) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H7B4, Q86TL8, Q8N5Z6, Q96AI5 | Gene names | SMYD3, ZMYND1, ZNFN3A1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.028826 (rank : 121) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TRI42_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.018760 (rank : 132) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IWZ5, Q8N832, Q8NDL3 | Gene names | TRIM42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.028359 (rank : 122) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.037786 (rank : 105) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CCD16_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.023255 (rank : 126) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R1N0, Q3TR52, Q9CWV9, Q9CYI6 | Gene names | Ccdc16, Omcg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16 (Ovus mutant candidate gene 1 protein). | |||||
|
CFDP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.021982 (rank : 130) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UEE9, O00393, O00404, Q9UEF0, Q9UEF1, Q9UEF8 | Gene names | CFDP1, BCNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur). | |||||
|
KRA57_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.073172 (rank : 45) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA98_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.044140 (rank : 95) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
MYSM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.032156 (rank : 111) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.004667 (rank : 153) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.031773 (rank : 113) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
STAB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.022195 (rank : 129) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
VEGFC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.028847 (rank : 120) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97953, Q543R6 | Gene names | Vegfc | |||
|
Domain Architecture |
|
|||||
| Description | Vascular endothelial growth factor C precursor (VEGF-C) (Vascular endothelial growth factor-related protein) (VRP) (Flt4 ligand) (Flt4- L). | |||||
|
ZNF31_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | -0.003063 (rank : 162) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17040, Q96H84 | Gene names | ZNF31, KOX29, ZNF360, ZSCAN20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 31 (Zinc finger protein KOX29) (Zinc finger and SCAN domain-containing protein 20) (Zinc finger protein 360). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.010531 (rank : 143) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
CSTN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.004211 (rank : 154) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ER65 | Gene names | Clstn2, Cs2, Cstn2 | |||
|
Domain Architecture |
|
|||||
| Description | Calsyntenin-2 precursor. | |||||
|
KR109_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.039637 (rank : 99) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
LUM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.002493 (rank : 157) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51884, Q96QM7 | Gene names | LUM, LDC | |||
|
Domain Architecture |

|
|||||
| Description | Lumican precursor (Keratan sulfate proteoglycan lumican) (KSPG lumican). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.022939 (rank : 127) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.036590 (rank : 107) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.068366 (rank : 49) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | -0.001705 (rank : 161) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.004077 (rank : 155) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.006496 (rank : 150) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.029947 (rank : 117) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
TAIP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.008069 (rank : 147) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59055 | Gene names | Taip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
TECT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.014465 (rank : 139) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q2MV57, Q6PDZ5, Q8C0B3, Q9CXF5 | Gene names | Tect2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tectonic-2 precursor. | |||||
|
ZNFX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.049953 (rank : 88) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
CC019_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.008757 (rank : 146) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PII3, Q96CS5 | Gene names | C3orf19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf19. | |||||
|
CDR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.029367 (rank : 119) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51861 | Gene names | CDR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related antigen 1 (CDR34). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.004018 (rank : 156) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CS1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.001669 (rank : 158) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.010102 (rank : 145) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
HTRA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.012086 (rank : 141) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
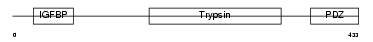
|
Domain Architecture |
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
HTRA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.011623 (rank : 142) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
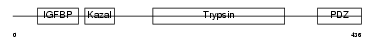
|
Domain Architecture |
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.010362 (rank : 144) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
KR10B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.042302 (rank : 97) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.030443 (rank : 115) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.029879 (rank : 118) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NP1L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.017137 (rank : 135) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55209 | Gene names | NAP1L1, NRP | |||
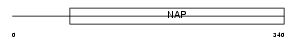
|
Domain Architecture |
|
|||||
| Description | Nucleosome assembly protein 1-like 1 (NAP-1-related protein) (hNRP). | |||||
|
SPY2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.016673 (rank : 136) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43597, Q5T6Z7 | Gene names | SPRY2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPY2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.017747 (rank : 133) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXV8, Q9WUQ9 | Gene names | Spry2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
TNR7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.025014 (rank : 125) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26842 | Gene names | TNFRSF7, CD27 | |||
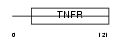
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27) (T14). | |||||
|
WNT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.004920 (rank : 152) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04628 | Gene names | WNT1, INT1 | |||
|
Domain Architecture |
|
|||||
| Description | Proto-oncogene protein Wnt-1 precursor. | |||||
|
WNT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.004921 (rank : 151) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04426 | Gene names | Wnt1, Int-1, Wnt-1 | |||
|
Domain Architecture |
|
|||||
| Description | Proto-oncogene protein Wnt-1 precursor. | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.063694 (rank : 60) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.066023 (rank : 52) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
CBX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.057697 (rank : 69) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.057697 (rank : 70) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.059231 (rank : 67) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.059817 (rank : 65) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CBX5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.054799 (rank : 74) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.054494 (rank : 78) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
DPF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.075289 (rank : 43) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
DPF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.066502 (rank : 50) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
K1333_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.064635 (rank : 57) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
K1333_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.064619 (rank : 58) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
LCE1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.050670 (rank : 83) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
LCE1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.063697 (rank : 59) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
LCE1E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.060890 (rank : 63) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.050454 (rank : 85) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.056384 (rank : 71) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.058437 (rank : 68) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.061172 (rank : 62) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.060008 (rank : 64) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.059267 (rank : 66) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.050036 (rank : 87) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.054688 (rank : 76) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.084830 (rank : 35) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.083252 (rank : 39) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.113410 (rank : 30) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
PHF12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.054632 (rank : 77) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.054708 (rank : 75) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.086731 (rank : 34) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
PHF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.084251 (rank : 38) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
PHF7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.070541 (rank : 47) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
PHF7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.065634 (rank : 55) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.078234 (rank : 41) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RAI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.084479 (rank : 37) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
REQU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.065921 (rank : 53) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.065455 (rank : 56) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.077877 (rank : 42) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.074775 (rank : 44) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
UHRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.052251 (rank : 81) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
UHRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.055055 (rank : 73) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
UHRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.050180 (rank : 86) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
EZH1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH1_MOUSE
|
||||||
| NC score | 0.998109 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | P70351, Q9R089 | Gene names | Ezh1, Enx2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH2_HUMAN
|
||||||
| NC score | 0.988594 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q15910, Q15755, Q92857 | Gene names | EZH2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
EZH2_MOUSE
|
||||||
| NC score | 0.988237 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q61188, Q9R090 | Gene names | Ezh2, Enx1h | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
SUV91_HUMAN
|
||||||
| NC score | 0.702841 (rank : 5) | θ value | 2.43343e-21 (rank : 5) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
SUV91_MOUSE
|
||||||
| NC score | 0.696594 (rank : 6) | θ value | 2.43343e-21 (rank : 6) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV92_HUMAN
|
||||||
| NC score | 0.691193 (rank : 7) | θ value | 7.82807e-20 (rank : 11) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV92_MOUSE
|
||||||
| NC score | 0.684997 (rank : 8) | θ value | 3.88503e-19 (rank : 15) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.651921 (rank : 9) | θ value | 7.82807e-20 (rank : 10) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.612885 (rank : 10) | θ value | 4.58923e-20 (rank : 8) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SETD8_HUMAN
|
||||||
| NC score | 0.611168 (rank : 11) | θ value | 1.47974e-10 (rank : 21) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
SETD8_MOUSE
|
||||||
| NC score | 0.610503 (rank : 12) | θ value | 1.47974e-10 (rank : 22) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q2YDW7, Q8C0J9 | Gene names | SETD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.564428 (rank : 13) | θ value | 5.99374e-20 (rank : 9) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.561693 (rank : 14) | θ value | 1.33526e-19 (rank : 12) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.555914 (rank : 15) | θ value | 3.17815e-21 (rank : 7) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.537792 (rank : 16) | θ value | 5.07402e-19 (rank : 16) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
SETB2_HUMAN
|
||||||
| NC score | 0.509735 (rank : 17) | θ value | 8.40245e-06 (rank : 25) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
SETB1_MOUSE
|
||||||
| NC score | 0.474638 (rank : 18) | θ value | 1.29631e-06 (rank : 23) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SETB1_HUMAN
|
||||||
| NC score | 0.469558 (rank : 19) | θ value | 2.21117e-06 (rank : 24) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15047, Q96GM9 | Gene names | SETDB1, KIAA0067 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.437011 (rank : 20) | θ value | 3.88503e-19 (rank : 14) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.422613 (rank : 21) | θ value | 1.13037e-18 (rank : 17) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.415198 (rank : 22) | θ value | 3.88503e-19 (rank : 13) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | 0.343227 (rank : 23) | θ value | 1.24977e-17 (rank : 19) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.341415 (rank : 24) | θ value | 5.60996e-18 (rank : 18) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EHMT1_HUMAN
|
||||||
| NC score | 0.334046 (rank : 25) | θ value | 2.7842e-17 (rank : 20) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
SETD7_MOUSE
|
||||||
| NC score | 0.207438 (rank : 26) | θ value | 0.00175202 (rank : 26) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VHL1 | Gene names | Setd7, Set7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7). | |||||
|
SETD7_HUMAN
|
||||||
| NC score | 0.201830 (rank : 27) | θ value | 0.00298849 (rank : 27) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WTS6, Q9C0E6 | Gene names | SETD7, KIAA1717, SET7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7) (Set9) (SET7/9). | |||||
|
MCSP_MOUSE
|
||||||
| NC score | 0.158880 (rank : 28) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.133721 (rank : 29) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PHF11_HUMAN
|
||||||
| NC score | 0.113410 (rank : 30) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.100161 (rank : 31) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.097708 (rank : 32) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.094522 (rank : 33) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
PHF6_HUMAN
|
||||||
| NC score | 0.086731 (rank : 34) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.084830 (rank : 35) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.084501 (rank : 36) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.084479 (rank : 37) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
PHF6_MOUSE
|
||||||
| NC score | 0.084251 (rank : 38) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.083252 (rank : 39) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
KRA24_HUMAN
|
||||||
| NC score | 0.082295 (rank : 40) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.078234 (rank : 41) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.077877 (rank : 42) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
DPF1_HUMAN
|
||||||
| NC score | 0.075289 (rank : 43) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.074775 (rank : 44) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
KRA57_HUMAN
|
||||||
| NC score | 0.073172 (rank : 45) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
SMYD3_HUMAN
|
||||||
| NC score | 0.070677 (rank : 46) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H7B4, Q86TL8, Q8N5Z6, Q96AI5 | Gene names | SMYD3, ZMYND1, ZNFN3A1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
PHF7_HUMAN
|
||||||
| NC score | 0.070541 (rank : 47) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
SMYD3_MOUSE
|
||||||
| NC score | 0.070181 (rank : 48) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CWR2, Q6P7V6, Q8BG90 | Gene names | Smyd3, Zmynd1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.068366 (rank : 49) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
DPF1_MOUSE
|
||||||
| NC score | 0.066502 (rank : 50) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.066334 (rank : 51) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.066023 (rank : 52) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
REQU_HUMAN
|
||||||
| NC score | 0.065921 (rank : 53) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
KRA44_HUMAN
|
||||||
| NC score | 0.065811 (rank : 54) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
PHF7_MOUSE
|
||||||
| NC score | 0.065634 (rank : 55) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
REQU_MOUSE
|
||||||
| NC score | 0.065455 (rank : 56) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
K1333_HUMAN
|
||||||
| NC score | 0.064635 (rank : 57) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
K1333_MOUSE
|
||||||
| NC score | 0.064619 (rank : 58) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
LCE1D_HUMAN
|
||||||
| NC score | 0.063697 (rank : 59) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.063694 (rank : 60) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
KRA45_HUMAN
|
||||||
| NC score | 0.061230 (rank : 61) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.061172 (rank : 62) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
LCE1E_HUMAN
|
||||||
| NC score | 0.060890 (rank : 63) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.060008 (rank : 64) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
CBX3_MOUSE
|
||||||
| NC score | 0.059817 (rank : 65) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.059267 (rank : 66) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
CBX3_HUMAN
|
||||||
| NC score | 0.059231 (rank : 67) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.058437 (rank : 68) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
CBX1_HUMAN
|
||||||
| NC score | 0.057697 (rank : 69) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| NC score | 0.057697 (rank : 70) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.056384 (rank : 71) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.055841 (rank : 72) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
UHRF1_MOUSE
|
||||||
| NC score | 0.055055 (rank : 73) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
CBX5_HUMAN
|
||||||
| NC score | 0.054799 (rank : 74) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.054708 (rank : 75) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.054688 (rank : 76) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.054632 (rank : 77) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
CBX5_MOUSE
|
||||||
| NC score | 0.054494 (rank : 78) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.053900 (rank : 79) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
KRA94_HUMAN
|
||||||
| NC score | 0.052886 (rank : 80) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BYQ2 | Gene names | KRTAP9-4, KAP9.4, KRTAP9.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-4 (Keratin-associated protein 9.4) (Ultrahigh sulfur keratin-associated protein 9.4). | |||||
|
UHRF1_HUMAN
|
||||||
| NC score | 0.052251 (rank : 81) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
ZNFX1_MOUSE
|
||||||
| NC score | 0.052236 (rank : 82) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
LCE1C_HUMAN
|
||||||
| NC score | 0.050670 (rank : 83) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
KR103_HUMAN
|
||||||
| NC score | 0.050630 (rank : 84) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.050454 (rank : 85) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
UHRF2_HUMAN
|
||||||
| NC score | 0.050180 (rank : 86) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.050036 (rank : 87) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
ZNFX1_HUMAN
|
||||||
| NC score | 0.049953 (rank : 88) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
KR102_HUMAN
|
||||||
| NC score | 0.048079 (rank : 89) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KRA93_HUMAN
|
||||||
| NC score | 0.047527 (rank : 90) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
TNR7_MOUSE
|
||||||
| NC score | 0.046799 (rank : 91) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P41272 | Gene names | Tnfrsf7, Cd27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.046678 (rank : 92) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.046268 (rank : 93) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.045559 (rank : 94) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
KRA98_HUMAN
|
||||||
| NC score | 0.044140 (rank : 95) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.042842 (rank : 96) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
KR10B_HUMAN
|
||||||
| NC score | 0.042302 (rank : 97) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.041878 (rank : 98) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
KR109_HUMAN
|
||||||
| NC score | 0.039637 (rank : 99) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.039512 (rank : 100) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CCD16_HUMAN
|
||||||
| NC score | 0.039458 (rank : 101) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96NB3, Q96F60, Q96GZ5, Q9BU38 | Gene names | CCDC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16. | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.038877 (rank : 102) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
ITB1_HUMAN
|
||||||
| NC score | 0.038706 (rank : 103) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P05556, P78466, P78467, Q13089, Q13090, Q13091, Q13212, Q14622, Q14647 | Gene names | ITGB1, FNRB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.038353 (rank : 104) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.037786 (rank : 105) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
ITB2_MOUSE
|
||||||
| NC score | 0.036853 (rank : 106) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P11835, Q64482 | Gene names | Itgb2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/P150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.036590 (rank : 107) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
SEPP1_HUMAN
|
||||||
| NC score | 0.035366 (rank : 108) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49908, Q6PD59, Q6PI43, Q6PI87, Q6PJF9 | Gene names | SEPP1, SELP | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein P precursor (SeP). | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.035191 (rank : 109) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
NOTC1_HUMAN
|
||||||
| NC score | 0.034805 (rank : 110) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
MYSM1_HUMAN
|
||||||
| NC score | 0.032156 (rank : 111) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.031960 (rank : 112) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.031773 (rank : 113) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
NP1L1_MOUSE
|
||||||
| NC score | 0.030665 (rank : 114) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28656, Q3UL14 | Gene names | Nap1l1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 1 (NAP-1-related protein) (Brain protein DN38). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.030443 (rank : 115) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
ITB2_HUMAN
|
||||||
| NC score | 0.030174 (rank : 116) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P05107, Q16418 | Gene names | ITGB2, CD18 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/p150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.029947 (rank : 117) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.029879 (rank : 118) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
CDR1_HUMAN
|
||||||
| NC score | 0.029367 (rank : 119) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51861 | Gene names | CDR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related antigen 1 (CDR34). | |||||
|
VEGFC_MOUSE
|
||||||
| NC score | 0.028847 (rank : 120) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97953, Q543R6 | Gene names | Vegfc | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor C precursor (VEGF-C) (Vascular endothelial growth factor-related protein) (VRP) (Flt4 ligand) (Flt4- L). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.028826 (rank : 121) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.028359 (rank : 122) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
PZRN3_MOUSE
|
||||||
| NC score | 0.026737 (rank : 123) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.025837 (rank : 124) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
TNR7_HUMAN
|
||||||
| NC score | 0.025014 (rank : 125) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26842 | Gene names | TNFRSF7, CD27 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27) (T14). | |||||
|
CCD16_MOUSE
|
||||||
| NC score | 0.023255 (rank : 126) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R1N0, Q3TR52, Q9CWV9, Q9CYI6 | Gene names | Ccdc16, Omcg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16 (Ovus mutant candidate gene 1 protein). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.022939 (rank : 127) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
PZRN3_HUMAN
|
||||||
| NC score | 0.022621 (rank : 128) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.022195 (rank : 129) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
CFDP1_HUMAN
|
||||||
| NC score | 0.021982 (rank : 130) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UEE9, O00393, O00404, Q9UEF0, Q9UEF1, Q9UEF8 | Gene names | CFDP1, BCNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur). | |||||
|
ACY3_HUMAN
|
||||||
| NC score | 0.021242 (rank : 131) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96HD9 | Gene names | ACY3, ASPA2 | |||
|
Domain Architecture |
|
|||||
| Description | Aspartoacylase-2 (EC 3.5.1.15) (Aminoacylase-3) (ACY-3) (Acylase III) (Hepatitis C virus core-binding protein 1) (HCBP1). | |||||
|
TRI42_HUMAN
|
||||||
| NC score | 0.018760 (rank : 132) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IWZ5, Q8N832, Q8NDL3 | Gene names | TRIM42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
SPY2_MOUSE
|
||||||
| NC score | 0.017747 (rank : 133) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXV8, Q9WUQ9 | Gene names | Spry2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
KIF4A_HUMAN
|
||||||
| NC score | 0.017720 (rank : 134) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
NP1L1_HUMAN
|
||||||
| NC score | 0.017137 (rank : 135) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55209 | Gene names | NAP1L1, NRP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 1 (NAP-1-related protein) (hNRP). | |||||
|
SPY2_HUMAN
|
||||||
| NC score | 0.016673 (rank : 136) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43597, Q5T6Z7 | Gene names | SPRY2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.016191 (rank : 137) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
NRDC_MOUSE
|
||||||
| NC score | 0.015735 (rank : 138) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
TECT2_MOUSE
|
||||||
| NC score | 0.014465 (rank : 139) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q2MV57, Q6PDZ5, Q8C0B3, Q9CXF5 | Gene names | Tect2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tectonic-2 precursor. | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.012783 (rank : 140) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
HTRA3_HUMAN
|
||||||
| NC score | 0.012086 (rank : 141) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
HTRA3_MOUSE
|
||||||
| NC score | 0.011623 (rank : 142) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.010531 (rank : 143) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.010362 (rank : 144) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.010102 (rank : 145) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
CC019_HUMAN
|
||||||
| NC score | 0.008757 (rank : 146) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PII3, Q96CS5 | Gene names | C3orf19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf19. | |||||
|
TAIP2_MOUSE
|
||||||
| NC score | 0.008069 (rank : 147) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59055 | Gene names | Taip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
TAF5_HUMAN
|
||||||
| NC score | 0.007854 (rank : 148) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15542, Q86UZ7, Q9Y4K5 | Gene names | TAF5, TAF2D | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 5 (Transcription initiation factor TFIID 100 kDa subunit) (TAF(II)100) (TAFII-100) (TAFII100). | |||||
|
TAF5_MOUSE
|
||||||
| NC score | 0.006624 (rank : 149) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C092, Q8BTR2 | Gene names | Taf5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 5 (Transcription initiation factor TFIID 100 kDa subunit) (TAF(II)100) (TAFII-100) (TAFII100). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.006496 (rank : 150) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
WNT1_MOUSE
|
||||||
| NC score | 0.004921 (rank : 151) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04426 | Gene names | Wnt1, Int-1, Wnt-1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein Wnt-1 precursor. | |||||
|
WNT1_HUMAN
|
||||||
| NC score | 0.004920 (rank : 152) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04628 | Gene names | WNT1, INT1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein Wnt-1 precursor. | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.004667 (rank : 153) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
CSTN2_MOUSE
|
||||||
| NC score | 0.004211 (rank : 154) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ER65 | Gene names | Clstn2, Cs2, Cstn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-2 precursor. | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.004077 (rank : 155) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.004018 (rank : 156) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
LUM_HUMAN
|
||||||
| NC score | 0.002493 (rank : 157) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51884, Q96QM7 | Gene names | LUM, LDC | |||
|
Domain Architecture |

|
|||||
| Description | Lumican precursor (Keratan sulfate proteoglycan lumican) (KSPG lumican). | |||||
|
CS1A_MOUSE
|
||||||
| NC score | 0.001669 (rank : 158) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
TRIO_HUMAN
|
||||||
| NC score | 0.001533 (rank : 159) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.000313 (rank : 160) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | -0.001705 (rank : 161) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
ZNF31_HUMAN
|
||||||
| NC score | -0.003063 (rank : 162) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17040, Q96H84 | Gene names | ZNF31, KOX29, ZNF360, ZSCAN20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 31 (Zinc finger protein KOX29) (Zinc finger and SCAN domain-containing protein 20) (Zinc finger protein 360). | |||||