Please be patient as the page loads
|
CBX1_MOUSE
|
||||||
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CBX1_HUMAN
|
||||||
| θ value | 9.41505e-74 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| θ value | 9.41505e-74 (rank : 2) | NC score | 0.999962 (rank : 2) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX5_MOUSE
|
||||||
| θ value | 1.7238e-59 (rank : 3) | NC score | 0.985998 (rank : 5) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX5_HUMAN
|
||||||
| θ value | 2.94036e-59 (rank : 4) | NC score | 0.984739 (rank : 6) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX3_MOUSE
|
||||||
| θ value | 5.02714e-51 (rank : 5) | NC score | 0.989971 (rank : 4) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CBX3_HUMAN
|
||||||
| θ value | 1.11993e-50 (rank : 6) | NC score | 0.990419 (rank : 3) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 7) | NC score | 0.630196 (rank : 14) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CBX2_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 8) | NC score | 0.672389 (rank : 9) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
SUV92_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 9) | NC score | 0.433804 (rank : 22) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
CBX4_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 10) | NC score | 0.639228 (rank : 10) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CBX4_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 11) | NC score | 0.634920 (rank : 12) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CBX7_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 12) | NC score | 0.695395 (rank : 8) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 13) | NC score | 0.271232 (rank : 26) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
CDYL1_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 14) | NC score | 0.491348 (rank : 17) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CBX7_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 15) | NC score | 0.716697 (rank : 7) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
SUV92_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 16) | NC score | 0.420643 (rank : 23) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
CDYL_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 17) | NC score | 0.479153 (rank : 19) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CBX8_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 18) | NC score | 0.634918 (rank : 13) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
CBX8_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 19) | NC score | 0.635133 (rank : 11) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QXV1 | Gene names | Cbx8, Pc3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (mPc3). | |||||
|
CDYL2_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 20) | NC score | 0.483161 (rank : 18) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
CBX6_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 21) | NC score | 0.614764 (rank : 16) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX6_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 22) | NC score | 0.622038 (rank : 15) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CDY1_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 23) | NC score | 0.455823 (rank : 21) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
CDY2_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 24) | NC score | 0.456279 (rank : 20) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
SUV91_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 25) | NC score | 0.399762 (rank : 24) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV91_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 26) | NC score | 0.380673 (rank : 25) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 27) | NC score | 0.189448 (rank : 27) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 28) | NC score | 0.188227 (rank : 28) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 29) | NC score | 0.179777 (rank : 29) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.099748 (rank : 38) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.098999 (rank : 40) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.124649 (rank : 32) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.101566 (rank : 37) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.116321 (rank : 33) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.114057 (rank : 34) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.113431 (rank : 35) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.099312 (rank : 39) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.023797 (rank : 84) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.002055 (rank : 90) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
A1AT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.007399 (rank : 88) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22599, Q61283, Q80ZH5, Q8VC20 | Gene names | Serpina1b, Aat2, Dom2, Spi1-2 | |||
|
Domain Architecture |
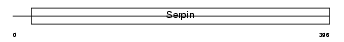
|
|||||
| Description | Alpha-1-antitrypsin 1-2 precursor (Serine protease inhibitor 1-2) (Alpha-1 protease inhibitor 2) (Alpha-1-antiproteinase) (AAT). | |||||
|
A1AT4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.007248 (rank : 89) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00897 | Gene names | Serpina1d, Dom4, Spi1-4 | |||
|
Domain Architecture |
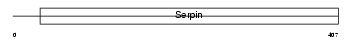
|
|||||
| Description | Alpha-1-antitrypsin 1-4 precursor (Serine protease inhibitor 1-4) (Alpha-1 protease inhibitor 4). | |||||
|
A1AT6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.007401 (rank : 87) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P81105, Q91V74, Q91WH5, Q91XC1 | Gene names | Spi1-6, Dom6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1-antitrypsin 1-6 precursor (Serine protease inhibitor 1-6) (Alpha-1 protease inhibitor 6). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.020218 (rank : 85) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
LACTB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.014849 (rank : 86) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P83111, P83096 | Gene names | LACTB | |||
|
Domain Architecture |
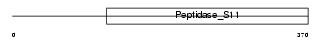
|
|||||
| Description | Serine beta-lactamase-like protein LACTB. | |||||
|
ATRX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.057827 (rank : 72) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
AUHM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.078654 (rank : 44) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLZ3, Q80YD7, Q8QZS0, Q9CY78, Q9D155 | Gene names | Auh | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding enoyl-CoA hydratase) (muAUH). | |||||
|
AUMH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.077710 (rank : 45) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13825, Q8WUE4 | Gene names | AUH | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding protein/enoyl-CoA hydratase). | |||||
|
BTAF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.063452 (rank : 57) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
ECH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.098729 (rank : 41) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13011, Q8WVX0, Q96EZ9 | Gene names | ECH1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.101733 (rank : 36) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35459, Q5M8P6 | Gene names | Ech1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECHA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.062442 (rank : 60) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40939, Q16679, Q96GT7 | Gene names | HADHA, HADH | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional enzyme subunit alpha, mitochondrial precursor (TP-alpha) (78 kDa gastrin-binding protein) [Includes: Long-chain enoyl-CoA hydratase (EC 4.2.1.17); Long chain 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.211)]. | |||||
|
ECHM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.096034 (rank : 43) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECHM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.096560 (rank : 42) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BH95, Q3TJK2, Q6GQS2, Q6PEN1, Q99LX7 | Gene names | Echs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECHP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.057947 (rank : 71) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBM2 | Gene names | Ehhadh | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
EP400_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.054834 (rank : 81) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
EP400_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.057698 (rank : 74) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
ERCC6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.065286 (rank : 53) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
EZH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.057697 (rank : 75) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.057323 (rank : 76) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70351, Q9R089 | Gene names | Ezh1, Enx2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.057709 (rank : 73) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15910, Q15755, Q92857 | Gene names | EZH2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
EZH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.056888 (rank : 77) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61188, Q9R090 | Gene names | Ezh2, Enx1h | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
INOC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.061077 (rank : 64) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
INOC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.061015 (rank : 65) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZPV2, Q6P7V0, Q6PCP1, Q8C9T7 | Gene names | Inoc1, Kiaa1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-). | |||||
|
MS3L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.055745 (rank : 79) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.059280 (rank : 70) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.059500 (rank : 69) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PECI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.147548 (rank : 30) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
PECI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.146675 (rank : 31) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
RA54B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.061680 (rank : 62) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y620 | Gene names | RAD54B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
RA54B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.061318 (rank : 63) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PFE3 | Gene names | Rad54b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
RAD54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.062801 (rank : 58) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92698, Q6IUY3 | Gene names | RAD54L, RAD54A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (hRAD54) (hHR54). | |||||
|
RAD54_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.062615 (rank : 59) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70270, Q8BSR5, Q8C2C4, Q8K3D4 | Gene names | Rad54l, Rad54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (mRAD54) (mHR54). | |||||
|
SETB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.063541 (rank : 56) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15047, Q96GM9 | Gene names | SETDB1, KIAA0067 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SETB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.062384 (rank : 61) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SETB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.060470 (rank : 68) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.067440 (rank : 50) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SETD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.060711 (rank : 66) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
SETD8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.060531 (rank : 67) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q2YDW7, Q8C0J9 | Gene names | SETD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.055056 (rank : 80) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
SMAL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.056163 (rank : 78) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
SMCA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.068168 (rank : 48) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28370, Q5JV41, Q5JV42 | Gene names | SMARCA1, SNF2L, SNF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1). | |||||
|
SMCA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.067941 (rank : 49) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.064667 (rank : 55) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.065394 (rank : 52) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SMCA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.068202 (rank : 46) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60264 | Gene names | SMARCA5, SNF2H, WCRF135 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin A5) (Sucrose nonfermenting protein 2 homolog) (hSNF2H). | |||||
|
SMCA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.068188 (rank : 47) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91ZW3, Q8C791, Q8CA22, Q8VDG1, Q925M9 | Gene names | Smarca5, Snf2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (Sucrose nonfermenting protein 2 homolog) (mSnf2h). | |||||
|
SMRCD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.065264 (rank : 54) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
|
SMRCD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.065627 (rank : 51) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q04692 | Gene names | Smarcad1, Etl1, Kiaa1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase SMARCAD1) (Enhancer trap locus homolog 1) (Etl-1). | |||||
|
TTF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.052074 (rank : 83) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.052720 (rank : 82) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
CBX1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 9.41505e-74 (rank : 1) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 2) | θ value | 9.41505e-74 (rank : 2) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX3_HUMAN
|
||||||
| NC score | 0.990419 (rank : 3) | θ value | 1.11993e-50 (rank : 6) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX3_MOUSE
|
||||||
| NC score | 0.989971 (rank : 4) | θ value | 5.02714e-51 (rank : 5) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CBX5_MOUSE
|
||||||
| NC score | 0.985998 (rank : 5) | θ value | 1.7238e-59 (rank : 3) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX5_HUMAN
|
||||||
| NC score | 0.984739 (rank : 6) | θ value | 2.94036e-59 (rank : 4) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX7_MOUSE
|
||||||
| NC score | 0.716697 (rank : 7) | θ value | 2.36244e-08 (rank : 15) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX7_HUMAN
|
||||||
| NC score | 0.695395 (rank : 8) | θ value | 1.06045e-08 (rank : 12) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX2_MOUSE
|
||||||
| NC score | 0.672389 (rank : 9) | θ value | 3.29651e-10 (rank : 8) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
CBX4_HUMAN
|
||||||
| NC score | 0.639228 (rank : 10) | θ value | 1.06045e-08 (rank : 10) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CBX8_MOUSE
|
||||||
| NC score | 0.635133 (rank : 11) | θ value | 1.53129e-07 (rank : 19) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QXV1 | Gene names | Cbx8, Pc3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (mPc3). | |||||
|
CBX4_MOUSE
|
||||||
| NC score | 0.634920 (rank : 12) | θ value | 1.06045e-08 (rank : 11) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CBX8_HUMAN
|
||||||
| NC score | 0.634918 (rank : 13) | θ value | 1.53129e-07 (rank : 18) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.630196 (rank : 14) | θ value | 3.29651e-10 (rank : 7) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CBX6_MOUSE
|
||||||
| NC score | 0.622038 (rank : 15) | θ value | 4.45536e-07 (rank : 22) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX6_HUMAN
|
||||||
| NC score | 0.614764 (rank : 16) | θ value | 4.45536e-07 (rank : 21) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CDYL1_HUMAN
|
||||||
| NC score | 0.491348 (rank : 17) | θ value | 1.80886e-08 (rank : 14) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL2_HUMAN
|
||||||
| NC score | 0.483161 (rank : 18) | θ value | 3.41135e-07 (rank : 20) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
CDYL_MOUSE
|
||||||
| NC score | 0.479153 (rank : 19) | θ value | 8.97725e-08 (rank : 17) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDY2_HUMAN
|
||||||
| NC score | 0.456279 (rank : 20) | θ value | 2.21117e-06 (rank : 24) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
CDY1_HUMAN
|
||||||
| NC score | 0.455823 (rank : 21) | θ value | 2.21117e-06 (rank : 23) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
SUV92_MOUSE
|
||||||
| NC score | 0.433804 (rank : 22) | θ value | 2.13673e-09 (rank : 9) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV92_HUMAN
|
||||||
| NC score | 0.420643 (rank : 23) | θ value | 5.26297e-08 (rank : 16) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV91_MOUSE
|
||||||
| NC score | 0.399762 (rank : 24) | θ value | 4.92598e-06 (rank : 25) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV91_HUMAN
|
||||||
| NC score | 0.380673 (rank : 25) | θ value | 1.09739e-05 (rank : 26) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.271232 (rank : 26) | θ value | 1.06045e-08 (rank : 13) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.189448 (rank : 27) | θ value | 0.00390308 (rank : 27) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.188227 (rank : 28) | θ value | 0.00665767 (rank : 28) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.179777 (rank : 29) | θ value | 0.0193708 (rank : 29) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
PECI_HUMAN
|
||||||
| NC score | 0.147548 (rank : 30) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
PECI_MOUSE
|
||||||
| NC score | 0.146675 (rank : 31) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.124649 (rank : 32) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.116321 (rank : 33) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.114057 (rank : 34) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.113431 (rank : 35) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
ECH1_MOUSE
|
||||||
| NC score | 0.101733 (rank : 36) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35459, Q5M8P6 | Gene names | Ech1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.101566 (rank : 37) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.099748 (rank : 38) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.099312 (rank : 39) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.098999 (rank : 40) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
ECH1_HUMAN
|
||||||
| NC score | 0.098729 (rank : 41) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13011, Q8WVX0, Q96EZ9 | Gene names | ECH1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECHM_MOUSE
|
||||||
| NC score | 0.096560 (rank : 42) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BH95, Q3TJK2, Q6GQS2, Q6PEN1, Q99LX7 | Gene names | Echs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECHM_HUMAN
|
||||||
| NC score | 0.096034 (rank : 43) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
AUHM_MOUSE
|
||||||
| NC score | 0.078654 (rank : 44) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLZ3, Q80YD7, Q8QZS0, Q9CY78, Q9D155 | Gene names | Auh | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding enoyl-CoA hydratase) (muAUH). | |||||
|
AUMH_HUMAN
|
||||||
| NC score | 0.077710 (rank : 45) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13825, Q8WUE4 | Gene names | AUH | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding protein/enoyl-CoA hydratase). | |||||
|
SMCA5_HUMAN
|
||||||
| NC score | 0.068202 (rank : 46) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60264 | Gene names | SMARCA5, SNF2H, WCRF135 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin A5) (Sucrose nonfermenting protein 2 homolog) (hSNF2H). | |||||
|
SMCA5_MOUSE
|
||||||
| NC score | 0.068188 (rank : 47) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91ZW3, Q8C791, Q8CA22, Q8VDG1, Q925M9 | Gene names | Smarca5, Snf2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (Sucrose nonfermenting protein 2 homolog) (mSnf2h). | |||||
|
SMCA1_HUMAN
|
||||||
| NC score | 0.068168 (rank : 48) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28370, Q5JV41, Q5JV42 | Gene names | SMARCA1, SNF2L, SNF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1). | |||||
|
SMCA1_MOUSE
|
||||||
| NC score | 0.067941 (rank : 49) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.067440 (rank : 50) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SMRCD_MOUSE
|
||||||
| NC score | 0.065627 (rank : 51) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q04692 | Gene names | Smarcad1, Etl1, Kiaa1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase SMARCAD1) (Enhancer trap locus homolog 1) (Etl-1). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.065394 (rank : 52) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
ERCC6_HUMAN
|
||||||
| NC score | 0.065286 (rank : 53) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
SMRCD_HUMAN
|
||||||
| NC score | 0.065264 (rank : 54) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.064667 (rank : 55) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SETB1_HUMAN
|
||||||
| NC score | 0.063541 (rank : 56) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15047, Q96GM9 | Gene names | SETDB1, KIAA0067 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
BTAF1_HUMAN
|
||||||
| NC score | 0.063452 (rank : 57) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
RAD54_HUMAN
|
||||||
| NC score | 0.062801 (rank : 58) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92698, Q6IUY3 | Gene names | RAD54L, RAD54A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (hRAD54) (hHR54). | |||||
|
RAD54_MOUSE
|
||||||
| NC score | 0.062615 (rank : 59) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70270, Q8BSR5, Q8C2C4, Q8K3D4 | Gene names | Rad54l, Rad54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (mRAD54) (mHR54). | |||||
|
ECHA_HUMAN
|
||||||
| NC score | 0.062442 (rank : 60) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40939, Q16679, Q96GT7 | Gene names | HADHA, HADH | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional enzyme subunit alpha, mitochondrial precursor (TP-alpha) (78 kDa gastrin-binding protein) [Includes: Long-chain enoyl-CoA hydratase (EC 4.2.1.17); Long chain 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.211)]. | |||||
|
SETB1_MOUSE
|
||||||
| NC score | 0.062384 (rank : 61) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
RA54B_HUMAN
|
||||||
| NC score | 0.061680 (rank : 62) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y620 | Gene names | RAD54B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
RA54B_MOUSE
|
||||||
| NC score | 0.061318 (rank : 63) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PFE3 | Gene names | Rad54b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
INOC1_HUMAN
|
||||||
| NC score | 0.061077 (rank : 64) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
INOC1_MOUSE
|
||||||
| NC score | 0.061015 (rank : 65) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZPV2, Q6P7V0, Q6PCP1, Q8C9T7 | Gene names | Inoc1, Kiaa1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-). | |||||
|
SETD8_HUMAN
|
||||||
| NC score | 0.060711 (rank : 66) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
SETD8_MOUSE
|
||||||
| NC score | 0.060531 (rank : 67) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q2YDW7, Q8C0J9 | Gene names | SETD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8). | |||||
|
SETB2_HUMAN
|
||||||
| NC score | 0.060470 (rank : 68) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.059500 (rank : 69) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.059280 (rank : 70) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
ECHP_MOUSE
|
||||||
| NC score | 0.057947 (rank : 71) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBM2 | Gene names | Ehhadh | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.057827 (rank : 72) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
EZH2_HUMAN
|
||||||
| NC score | 0.057709 (rank : 73) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15910, Q15755, Q92857 | Gene names | EZH2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.057698 (rank : 74) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
EZH1_HUMAN
|
||||||
| NC score | 0.057697 (rank : 75) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH1_MOUSE
|
||||||
| NC score | 0.057323 (rank : 76) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70351, Q9R089 | Gene names | Ezh1, Enx2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH2_MOUSE
|
||||||
| NC score | 0.056888 (rank : 77) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61188, Q9R090 | Gene names | Ezh2, Enx1h | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
SMAL1_MOUSE
|
||||||
| NC score | 0.056163 (rank : 78) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
MS3L1_MOUSE
|
||||||
| NC score | 0.055745 (rank : 79) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.055056 (rank : 80) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.054834 (rank : 81) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.052720 (rank : 82) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
TTF2_HUMAN
|
||||||
| NC score | 0.052074 (rank : 83) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.023797 (rank : 84) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.020218 (rank : 85) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
LACTB_HUMAN
|
||||||
| NC score | 0.014849 (rank : 86) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P83111, P83096 | Gene names | LACTB | |||
|
Domain Architecture |

|
|||||
| Description | Serine beta-lactamase-like protein LACTB. | |||||
|
A1AT6_MOUSE
|
||||||
| NC score | 0.007401 (rank : 87) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P81105, Q91V74, Q91WH5, Q91XC1 | Gene names | Spi1-6, Dom6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1-antitrypsin 1-6 precursor (Serine protease inhibitor 1-6) (Alpha-1 protease inhibitor 6). | |||||
|
A1AT2_MOUSE
|
||||||
| NC score | 0.007399 (rank : 88) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22599, Q61283, Q80ZH5, Q8VC20 | Gene names | Serpina1b, Aat2, Dom2, Spi1-2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antitrypsin 1-2 precursor (Serine protease inhibitor 1-2) (Alpha-1 protease inhibitor 2) (Alpha-1-antiproteinase) (AAT). | |||||
|
A1AT4_MOUSE
|
||||||
| NC score | 0.007248 (rank : 89) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00897 | Gene names | Serpina1d, Dom4, Spi1-4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antitrypsin 1-4 precursor (Serine protease inhibitor 1-4) (Alpha-1 protease inhibitor 4). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.002055 (rank : 90) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||