Please be patient as the page loads
|
CBX6_HUMAN
|
||||||
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CBX6_MOUSE
|
||||||
| θ value | 4.12933e-154 (rank : 1) | NC score | 0.976218 (rank : 2) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX6_HUMAN
|
||||||
| θ value | 5.05951e-144 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX8_HUMAN
|
||||||
| θ value | 6.17384e-33 (rank : 3) | NC score | 0.880551 (rank : 6) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
CBX8_MOUSE
|
||||||
| θ value | 6.17384e-33 (rank : 4) | NC score | 0.882969 (rank : 5) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QXV1 | Gene names | Cbx8, Pc3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (mPc3). | |||||
|
CBX4_HUMAN
|
||||||
| θ value | 2.42779e-29 (rank : 5) | NC score | 0.846348 (rank : 7) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CBX4_MOUSE
|
||||||
| θ value | 1.02001e-27 (rank : 6) | NC score | 0.840305 (rank : 8) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CBX7_HUMAN
|
||||||
| θ value | 2.96777e-27 (rank : 7) | NC score | 0.889845 (rank : 3) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX7_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 8) | NC score | 0.889670 (rank : 4) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 5.42112e-21 (rank : 9) | NC score | 0.759134 (rank : 10) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CBX2_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 10) | NC score | 0.798726 (rank : 9) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
CBX5_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 11) | NC score | 0.614326 (rank : 13) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX5_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 12) | NC score | 0.613356 (rank : 14) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 13) | NC score | 0.614764 (rank : 11) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 14) | NC score | 0.614764 (rank : 12) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CDYL_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 15) | NC score | 0.391521 (rank : 17) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CBX3_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 16) | NC score | 0.585001 (rank : 15) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX3_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 17) | NC score | 0.572523 (rank : 16) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CDYL1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 18) | NC score | 0.382632 (rank : 18) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDY1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 19) | NC score | 0.365829 (rank : 21) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
CDY2_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 20) | NC score | 0.366627 (rank : 20) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 21) | NC score | 0.211236 (rank : 26) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
SUV91_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 22) | NC score | 0.292324 (rank : 25) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
CDYL2_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 23) | NC score | 0.380571 (rank : 19) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 24) | NC score | 0.071386 (rank : 37) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SUV91_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 25) | NC score | 0.309868 (rank : 23) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV92_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 26) | NC score | 0.323154 (rank : 22) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 27) | NC score | 0.159187 (rank : 29) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 28) | NC score | 0.165795 (rank : 27) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 29) | NC score | 0.164302 (rank : 28) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 30) | NC score | 0.059991 (rank : 56) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 31) | NC score | 0.056978 (rank : 67) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SUV92_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 32) | NC score | 0.299620 (rank : 24) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
PRD16_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 33) | NC score | 0.006825 (rank : 144) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HAZ2, Q8WYJ9, Q9C0I8 | Gene names | PRDM16, KIAA1675, MEL1, PFM13 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 16 (PR domain-containing protein 16) (Transcription factor MEL1). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 34) | NC score | 0.033949 (rank : 113) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 35) | NC score | 0.043235 (rank : 98) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 36) | NC score | 0.040906 (rank : 104) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 37) | NC score | 0.053663 (rank : 83) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
K1914_MOUSE
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.058950 (rank : 58) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5DTU0, Q8BID1, Q8K2G0 | Gene names | Kiaa1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
AKAP2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.058229 (rank : 63) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.043134 (rank : 99) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.059767 (rank : 57) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.024249 (rank : 118) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.020267 (rank : 124) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
AKAP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.045328 (rank : 96) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
DACT1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.041166 (rank : 103) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
LIMD1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.018069 (rank : 128) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.038357 (rank : 106) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PGCB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.023909 (rank : 119) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.044883 (rank : 97) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.038164 (rank : 107) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.039793 (rank : 105) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.021169 (rank : 122) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.009074 (rank : 140) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
PGCB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.019006 (rank : 126) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.036009 (rank : 111) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
EGR4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.003912 (rank : 149) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.031971 (rank : 114) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.051001 (rank : 89) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
C1QR1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.005846 (rank : 148) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NPY3, O00274 | Gene names | CD93, C1QR1, MXRA4 | |||
|
Domain Architecture |
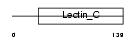
|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (CDw93). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.013272 (rank : 134) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
DNM3A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.022036 (rank : 121) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.037894 (rank : 108) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EXTL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.014956 (rank : 133) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JKV7 | Gene names | Extl1 | |||
|
Domain Architecture |

|
|||||
| Description | Exostosin-like 1 (EC 2.4.1.224) (Glucuronosyl-N-acetylglucosaminyl- proteoglycan 4-alpha-N-acetylglucosaminyltransferase) (Exostosin-L) (Multiple exostosis-like protein). | |||||
|
K1914_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.046168 (rank : 94) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N4X5, Q8TB54, Q96PX4, Q96SY5 | Gene names | KIAA1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.036985 (rank : 110) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.034566 (rank : 112) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.082815 (rank : 32) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.029901 (rank : 116) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
TESK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | -0.000300 (rank : 152) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96S53, Q8N520, Q9Y3Q6 | Gene names | TESK2 | |||
|
Domain Architecture |
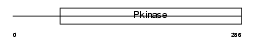
|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
TNR4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.017328 (rank : 130) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43489, Q13663, Q5T7M0 | Gene names | TNFRSF4, TXGP1L | |||
|
Domain Architecture |
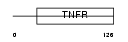
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (ACT35 antigen) (TAX transcriptionally-activated glycoprotein 1 receptor) (CD134 antigen). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.048232 (rank : 91) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.046126 (rank : 95) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
DCX_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.012004 (rank : 137) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.006228 (rank : 147) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
K1849_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.012698 (rank : 135) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.046947 (rank : 92) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SON_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.030492 (rank : 115) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
T53I2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.020096 (rank : 125) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFU8, Q711P5, Q8CHM4 | Gene names | Tp53inp2, Dor, Trp53inp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p53-inducible nuclear protein 2 (Diabetes and obesity- regulated protein). | |||||
|
ULK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.000380 (rank : 150) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.057908 (rank : 64) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.058658 (rank : 61) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
CP007_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.016968 (rank : 132) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
DZIP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.009936 (rank : 139) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86YF9, Q8WY45, Q8WY46, Q9UGA5, Q9Y2K0 | Gene names | DZIP1, DZIP, DZIP2, KIAA0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1/2). | |||||
|
HAIR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.012248 (rank : 136) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.042988 (rank : 100) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.011711 (rank : 138) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
TCF19_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.028317 (rank : 117) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.017246 (rank : 131) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
KI2S3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.006437 (rank : 145) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14952, O00644 | Gene names | KIR2DS3, NKAT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS3 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 7) (NKAT-7). | |||||
|
KI3L2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.006335 (rank : 146) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43630, Q13238, Q14947, Q14948, Q92684, Q95367 | Gene names | KIR3DL2, CD158K, NKAT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL2 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 4) (NKAT-4) (p70 natural killer cell receptor clone CL-5) (CD158k antigen). | |||||
|
PHC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.020779 (rank : 123) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P78364, Q8WVM3, Q9BU63 | Gene names | PHC1, EDR1, PH1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (hPH1) (Early development regulatory protein 1). | |||||
|
ULK2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.000028 (rank : 151) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QY01, Q80TV7, Q9WTP4 | Gene names | Ulk2, Kiaa0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2) (Serine/threonine-protein kinase Unc51.2). | |||||
|
ZHX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.007759 (rank : 143) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.022389 (rank : 120) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.046439 (rank : 93) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.037406 (rank : 109) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
BTBD7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.008218 (rank : 142) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P203, Q69Z05, Q7Z308, Q86TS0, Q9HAA4, Q9NVM0 | Gene names | BTBD7, KIAA1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.068180 (rank : 44) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.070544 (rank : 40) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
GA2L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.017757 (rank : 129) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.042052 (rank : 101) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.041675 (rank : 102) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RPGF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.009010 (rank : 141) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
T240L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.018120 (rank : 127) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6JPI3, Q3TRF4, Q3UQI8, Q80TM3, Q80WQ0 | Gene names | Thrap2, Kiaa1025, Trap240l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
AUHM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.056026 (rank : 69) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLZ3, Q80YD7, Q8QZS0, Q9CY78, Q9D155 | Gene names | Auh | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding enoyl-CoA hydratase) (muAUH). | |||||
|
AUMH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.055376 (rank : 71) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13825, Q8WUE4 | Gene names | AUH | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding protein/enoyl-CoA hydratase). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.054426 (rank : 76) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.071245 (rank : 38) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.061836 (rank : 53) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.061871 (rank : 51) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.058849 (rank : 59) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.054311 (rank : 79) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.053074 (rank : 85) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.056083 (rank : 68) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.053699 (rank : 82) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.057114 (rank : 66) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.057214 (rank : 65) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
ECH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.070500 (rank : 41) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13011, Q8WVX0, Q96EZ9 | Gene names | ECH1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.072630 (rank : 36) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35459, Q5M8P6 | Gene names | Ech1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECHM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.068505 (rank : 43) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECHM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.068910 (rank : 42) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BH95, Q3TJK2, Q6GQS2, Q6PEN1, Q99LX7 | Gene names | Echs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
EP400_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.054588 (rank : 75) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
EP400_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.055368 (rank : 73) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
GGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.055008 (rank : 74) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.058779 (rank : 60) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.052060 (rank : 87) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.055444 (rank : 70) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.054418 (rank : 77) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.062703 (rank : 49) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MS3L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.074324 (rank : 35) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.078873 (rank : 34) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PECI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.105427 (rank : 30) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
PECI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.104822 (rank : 31) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.062723 (rank : 48) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.062600 (rank : 50) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.058597 (rank : 62) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.067276 (rank : 46) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.079265 (rank : 33) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.061852 (rank : 52) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.054321 (rank : 78) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SET1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.067378 (rank : 45) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.052213 (rank : 86) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.055376 (rank : 72) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.053607 (rank : 84) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.061281 (rank : 55) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.053856 (rank : 80) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.063319 (rank : 47) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.070660 (rank : 39) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.061359 (rank : 54) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.050128 (rank : 90) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.053727 (rank : 81) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.051188 (rank : 88) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
CBX6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 5.05951e-144 (rank : 2) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX6_MOUSE
|
||||||
| NC score | 0.976218 (rank : 2) | θ value | 4.12933e-154 (rank : 1) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX7_HUMAN
|
||||||
| NC score | 0.889845 (rank : 3) | θ value | 2.96777e-27 (rank : 7) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX7_MOUSE
|
||||||
| NC score | 0.889670 (rank : 4) | θ value | 8.36355e-22 (rank : 8) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX8_MOUSE
|
||||||
| NC score | 0.882969 (rank : 5) | θ value | 6.17384e-33 (rank : 4) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QXV1 | Gene names | Cbx8, Pc3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (mPc3). | |||||
|
CBX8_HUMAN
|
||||||
| NC score | 0.880551 (rank : 6) | θ value | 6.17384e-33 (rank : 3) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
CBX4_HUMAN
|
||||||
| NC score | 0.846348 (rank : 7) | θ value | 2.42779e-29 (rank : 5) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CBX4_MOUSE
|
||||||
| NC score | 0.840305 (rank : 8) | θ value | 1.02001e-27 (rank : 6) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CBX2_MOUSE
|
||||||
| NC score | 0.798726 (rank : 9) | θ value | 5.42112e-21 (rank : 10) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.759134 (rank : 10) | θ value | 5.42112e-21 (rank : 9) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CBX1_HUMAN
|
||||||
| NC score | 0.614764 (rank : 11) | θ value | 4.45536e-07 (rank : 13) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| NC score | 0.614764 (rank : 12) | θ value | 4.45536e-07 (rank : 14) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX5_MOUSE
|
||||||
| NC score | 0.614326 (rank : 13) | θ value | 3.08544e-08 (rank : 11) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX5_HUMAN
|
||||||
| NC score | 0.613356 (rank : 14) | θ value | 4.0297e-08 (rank : 12) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX3_HUMAN
|
||||||
| NC score | 0.585001 (rank : 15) | θ value | 2.44474e-05 (rank : 16) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX3_MOUSE
|
||||||
| NC score | 0.572523 (rank : 16) | θ value | 0.000121331 (rank : 17) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CDYL_MOUSE
|
||||||
| NC score | 0.391521 (rank : 17) | θ value | 6.43352e-06 (rank : 15) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL1_HUMAN
|
||||||
| NC score | 0.382632 (rank : 18) | θ value | 0.000121331 (rank : 18) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL2_HUMAN
|
||||||
| NC score | 0.380571 (rank : 19) | θ value | 0.00035302 (rank : 23) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
CDY2_HUMAN
|
||||||
| NC score | 0.366627 (rank : 20) | θ value | 0.00020696 (rank : 20) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
CDY1_HUMAN
|
||||||
| NC score | 0.365829 (rank : 21) | θ value | 0.00020696 (rank : 19) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
SUV92_MOUSE
|
||||||
| NC score | 0.323154 (rank : 22) | θ value | 0.000602161 (rank : 26) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV91_MOUSE
|
||||||
| NC score | 0.309868 (rank : 23) | θ value | 0.000461057 (rank : 25) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV92_HUMAN
|
||||||
| NC score | 0.299620 (rank : 24) | θ value | 0.0148317 (rank : 32) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV91_HUMAN
|
||||||
| NC score | 0.292324 (rank : 25) | θ value | 0.00020696 (rank : 22) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.211236 (rank : 26) | θ value | 0.00020696 (rank : 21) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.165795 (rank : 27) | θ value | 0.00175202 (rank : 28) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.164302 (rank : 28) | θ value | 0.00175202 (rank : 29) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.159187 (rank : 29) | θ value | 0.00102713 (rank : 27) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
PECI_HUMAN
|
||||||
| NC score | 0.105427 (rank : 30) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
PECI_MOUSE
|
||||||
| NC score | 0.104822 (rank : 31) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.082815 (rank : 32) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.079265 (rank : 33) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.078873 (rank : 34) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MS3L1_MOUSE
|
||||||
| NC score | 0.074324 (rank : 35) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
ECH1_MOUSE
|
||||||
| NC score | 0.072630 (rank : 36) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35459, Q5M8P6 | Gene names | Ech1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.071386 (rank : 37) | θ value | 0.00035302 (rank : 24) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.071245 (rank : 38) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.070660 (rank : 39) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.070544 (rank : 40) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
ECH1_HUMAN
|
||||||
| NC score | 0.070500 (rank : 41) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13011, Q8WVX0, Q96EZ9 | Gene names | ECH1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECHM_MOUSE
|
||||||
| NC score | 0.068910 (rank : 42) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BH95, Q3TJK2, Q6GQS2, Q6PEN1, Q99LX7 | Gene names | Echs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECHM_HUMAN
|
||||||
| NC score | 0.068505 (rank : 43) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.068180 (rank : 44) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.067378 (rank : 45) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.067276 (rank : 46) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.063319 (rank : 47) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.062723 (rank : 48) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.062703 (rank : 49) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.062600 (rank : 50) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.061871 (rank : 51) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.061852 (rank : 52) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.061836 (rank : 53) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.061359 (rank : 54) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.061281 (rank : 55) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.059991 (rank : 56) | θ value | 0.0113563 (rank : 30) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.059767 (rank : 57) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
K1914_MOUSE
|
||||||
| NC score | 0.058950 (rank : 58) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5DTU0, Q8BID1, Q8K2G0 | Gene names | Kiaa1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.058849 (rank : 59) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.058779 (rank : 60) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.058658 (rank : 61) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.058597 (rank : 62) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
AKAP2_MOUSE
|
||||||
| NC score | 0.058229 (rank : 63) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.057908 (rank : 64) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.057214 (rank : 65) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.057114 (rank : 66) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.056978 (rank : 67) | θ value | 0.0148317 (rank : 31) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.056083 (rank : 68) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
AUHM_MOUSE
|
||||||
| NC score | 0.056026 (rank : 69) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLZ3, Q80YD7, Q8QZS0, Q9CY78, Q9D155 | Gene names | Auh | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding enoyl-CoA hydratase) (muAUH). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.055444 (rank : 70) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
AUMH_HUMAN
|
||||||
| NC score | 0.055376 (rank : 71) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13825, Q8WUE4 | Gene names | AUH | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding protein/enoyl-CoA hydratase). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.055376 (rank : 72) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.055368 (rank : 73) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.055008 (rank : 74) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.054588 (rank : 75) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.054426 (rank : 76) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.054418 (rank : 77) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.054321 (rank : 78) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.054311 (rank : 79) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.053856 (rank : 80) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.053727 (rank : 81) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.053699 (rank : 82) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.053663 (rank : 83) | θ value | 0.0961366 (rank : 37) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.053607 (rank : 84) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.053074 (rank : 85) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.052213 (rank : 86) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.052060 (rank : 87) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.051188 (rank : 88) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.051001 (rank : 89) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.050128 (rank : 90) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.048232 (rank : 91) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.046947 (rank : 92) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.046439 (rank : 93) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
K1914_HUMAN
|
||||||
| NC score | 0.046168 (rank : 94) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N4X5, Q8TB54, Q96PX4, Q96SY5 | Gene names | KIAA1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.046126 (rank : 95) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
AKAP2_HUMAN
|
||||||
| NC score | 0.045328 (rank : 96) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.044883 (rank : 97) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.043235 (rank : 98) | θ value | 0.0961366 (rank : 35) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.043134 (rank : 99) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.042988 (rank : 100) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.042052 (rank : 101) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.041675 (rank : 102) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
DACT1_HUMAN
|
||||||
| NC score | 0.041166 (rank : 103) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.040906 (rank : 104) | θ value | 0.0961366 (rank : 36) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.039793 (rank : 105) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.038357 (rank : 106) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.038164 (rank : 107) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.037894 (rank : 108) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.037406 (rank : 109) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.036985 (rank : 110) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.036009 (rank : 111) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.034566 (rank : 112) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.033949 (rank : 113) | θ value | 0.0330416 (rank : 34) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.031971 (rank : 114) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.030492 (rank : 115) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.029901 (rank : 116) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
TCF19_HUMAN
|
||||||
| NC score | 0.028317 (rank : 117) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.024249 (rank : 118) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
PGCB_HUMAN
|
||||||
| NC score | 0.023909 (rank : 119) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.022389 (rank : 120) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
DNM3A_HUMAN
|
||||||
| NC score | 0.022036 (rank : 121) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.021169 (rank : 122) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
PHC1_HUMAN
|
||||||
| NC score | 0.020779 (rank : 123) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P78364, Q8WVM3, Q9BU63 | Gene names | PHC1, EDR1, PH1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (hPH1) (Early development regulatory protein 1). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.020267 (rank : 124) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
T53I2_MOUSE
|
||||||
| NC score | 0.020096 (rank : 125) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFU8, Q711P5, Q8CHM4 | Gene names | Tp53inp2, Dor, Trp53inp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p53-inducible nuclear protein 2 (Diabetes and obesity- regulated protein). | |||||
|
PGCB_MOUSE
|
||||||
| NC score | 0.019006 (rank : 126) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
T240L_MOUSE
|
||||||
| NC score | 0.018120 (rank : 127) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6JPI3, Q3TRF4, Q3UQI8, Q80TM3, Q80WQ0 | Gene names | Thrap2, Kiaa1025, Trap240l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
LIMD1_MOUSE
|
||||||
| NC score | 0.018069 (rank : 128) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
GA2L1_HUMAN
|
||||||
| NC score | 0.017757 (rank : 129) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
TNR4_HUMAN
|
||||||
| NC score | 0.017328 (rank : 130) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43489, Q13663, Q5T7M0 | Gene names | TNFRSF4, TXGP1L | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (ACT35 antigen) (TAX transcriptionally-activated glycoprotein 1 receptor) (CD134 antigen). | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.017246 (rank : 131) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
CP007_MOUSE
|
||||||
| NC score | 0.016968 (rank : 132) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
EXTL1_MOUSE
|
||||||
| NC score | 0.014956 (rank : 133) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JKV7 | Gene names | Extl1 | |||
|
Domain Architecture |

|
|||||
| Description | Exostosin-like 1 (EC 2.4.1.224) (Glucuronosyl-N-acetylglucosaminyl- proteoglycan 4-alpha-N-acetylglucosaminyltransferase) (Exostosin-L) (Multiple exostosis-like protein). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.013272 (rank : 134) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
K1849_MOUSE
|
||||||
| NC score | 0.012698 (rank : 135) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
HAIR_HUMAN
|
||||||
| NC score | 0.012248 (rank : 136) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
DCX_MOUSE
|
||||||
| NC score | 0.012004 (rank : 137) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.011711 (rank : 138) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
DZIP1_HUMAN
|
||||||
| NC score | 0.009936 (rank : 139) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86YF9, Q8WY45, Q8WY46, Q9UGA5, Q9Y2K0 | Gene names | DZIP1, DZIP, DZIP2, KIAA0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1/2). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.009074 (rank : 140) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
RPGF1_HUMAN
|
||||||
| NC score | 0.009010 (rank : 141) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
BTBD7_HUMAN
|
||||||
| NC score | 0.008218 (rank : 142) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P203, Q69Z05, Q7Z308, Q86TS0, Q9HAA4, Q9NVM0 | Gene names | BTBD7, KIAA1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
ZHX1_MOUSE
|
||||||
| NC score | 0.007759 (rank : 143) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
PRD16_HUMAN
|
||||||
| NC score | 0.006825 (rank : 144) | θ value | 0.0193708 (rank : 33) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HAZ2, Q8WYJ9, Q9C0I8 | Gene names | PRDM16, KIAA1675, MEL1, PFM13 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 16 (PR domain-containing protein 16) (Transcription factor MEL1). | |||||
|
KI2S3_HUMAN
|
||||||
| NC score | 0.006437 (rank : 145) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14952, O00644 | Gene names | KIR2DS3, NKAT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS3 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 7) (NKAT-7). | |||||
|
KI3L2_HUMAN
|
||||||
| NC score | 0.006335 (rank : 146) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43630, Q13238, Q14947, Q14948, Q92684, Q95367 | Gene names | KIR3DL2, CD158K, NKAT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL2 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 4) (NKAT-4) (p70 natural killer cell receptor clone CL-5) (CD158k antigen). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.006228 (rank : 147) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
C1QR1_HUMAN
|
||||||
| NC score | 0.005846 (rank : 148) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NPY3, O00274 | Gene names | CD93, C1QR1, MXRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (CDw93). | |||||
|
EGR4_HUMAN
|
||||||
| NC score | 0.003912 (rank : 149) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
ULK2_HUMAN
|
||||||
| NC score | 0.000380 (rank : 150) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
ULK2_MOUSE
|
||||||
| NC score | 0.000028 (rank : 151) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QY01, Q80TV7, Q9WTP4 | Gene names | Ulk2, Kiaa0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2) (Serine/threonine-protein kinase Unc51.2). | |||||
|
TESK2_HUMAN
|
||||||
| NC score | -0.000300 (rank : 152) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96S53, Q8N520, Q9Y3Q6 | Gene names | TESK2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||