Please be patient as the page loads
|
SUV92_HUMAN
|
||||||
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SUV92_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV92_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998116 (rank : 2) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV91_MOUSE
|
||||||
| θ value | 4.14853e-138 (rank : 3) | NC score | 0.986798 (rank : 4) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV91_HUMAN
|
||||||
| θ value | 9.24197e-138 (rank : 4) | NC score | 0.986825 (rank : 3) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | 1.08726e-37 (rank : 5) | NC score | 0.418616 (rank : 24) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 1.42001e-37 (rank : 6) | NC score | 0.415044 (rank : 25) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EHMT1_HUMAN
|
||||||
| θ value | 3.4976e-36 (rank : 7) | NC score | 0.408810 (rank : 28) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 8) | NC score | 0.591450 (rank : 15) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 8.93572e-24 (rank : 9) | NC score | 0.589334 (rank : 16) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 10) | NC score | 0.681190 (rank : 9) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
EZH1_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 11) | NC score | 0.691193 (rank : 6) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH1_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 12) | NC score | 0.690049 (rank : 7) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70351, Q9R089 | Gene names | Ezh1, Enx2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH2_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 13) | NC score | 0.691368 (rank : 5) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15910, Q15755, Q92857 | Gene names | EZH2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
EZH2_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 14) | NC score | 0.686122 (rank : 8) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61188, Q9R090 | Gene names | Ezh2, Enx1h | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 15) | NC score | 0.539451 (rank : 18) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 16) | NC score | 0.520446 (rank : 19) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
SETB1_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 17) | NC score | 0.625135 (rank : 12) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15047, Q96GM9 | Gene names | SETDB1, KIAA0067 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SETB1_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 18) | NC score | 0.627124 (rank : 11) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 19) | NC score | 0.398949 (rank : 30) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 20) | NC score | 0.401426 (rank : 29) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 21) | NC score | 0.572959 (rank : 17) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 22) | NC score | 0.381437 (rank : 31) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
SETB2_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 23) | NC score | 0.636978 (rank : 10) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
SETD8_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 24) | NC score | 0.607985 (rank : 13) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
SETD8_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 25) | NC score | 0.607620 (rank : 14) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q2YDW7, Q8C0J9 | Gene names | SETD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8). | |||||
|
CBX3_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 26) | NC score | 0.422798 (rank : 20) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CBX3_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 27) | NC score | 0.422365 (rank : 21) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX5_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 28) | NC score | 0.410837 (rank : 26) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX5_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 29) | NC score | 0.409976 (rank : 27) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX1_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 30) | NC score | 0.420643 (rank : 22) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 31) | NC score | 0.420643 (rank : 23) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX4_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 32) | NC score | 0.314991 (rank : 35) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CBX4_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 33) | NC score | 0.312888 (rank : 36) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 34) | NC score | 0.308278 (rank : 38) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CBX2_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 35) | NC score | 0.327677 (rank : 34) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
CBX7_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 36) | NC score | 0.338432 (rank : 33) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX7_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 37) | NC score | 0.355095 (rank : 32) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX8_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 38) | NC score | 0.309653 (rank : 37) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
CBX8_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 39) | NC score | 0.305555 (rank : 39) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QXV1 | Gene names | Cbx8, Pc3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (mPc3). | |||||
|
CDY1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 40) | NC score | 0.205995 (rank : 43) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
CDY2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 41) | NC score | 0.205228 (rank : 44) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
CDYL2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 42) | NC score | 0.218735 (rank : 42) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 43) | NC score | 0.168535 (rank : 47) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
CBX6_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 44) | NC score | 0.299620 (rank : 40) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX6_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 45) | NC score | 0.299209 (rank : 41) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 46) | NC score | 0.069522 (rank : 62) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
CDYL_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 47) | NC score | 0.201739 (rank : 46) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 48) | NC score | 0.203173 (rank : 45) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 49) | NC score | 0.069871 (rank : 60) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 50) | NC score | 0.069665 (rank : 61) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
SMYD3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 51) | NC score | 0.091999 (rank : 52) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CWR2, Q6P7V6, Q8BG90 | Gene names | Smyd3, Zmynd1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
SMYD3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.092520 (rank : 51) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H7B4, Q86TL8, Q8N5Z6, Q96AI5 | Gene names | SMYD3, ZMYND1, ZNFN3A1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
IFNA4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.010379 (rank : 101) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07351 | Gene names | Ifna4 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon alpha-4 precursor. | |||||
|
SO1B3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.005346 (rank : 102) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPD5 | Gene names | SLCO1B3, LST2, SLC21A8 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 1B3 (Solute carrier family 21 member 8) (Organic anion transporter 8) (Organic anion-transporting polypeptide 8) (OATP8) (Liver-specific organic anion transporter 2) (LST-2). | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.064312 (rank : 68) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.064300 (rank : 69) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
ANKR6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.051330 (rank : 95) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
ANKY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.053153 (rank : 85) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IV38, Q96BL3 | Gene names | ANKMY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
ANKY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.052270 (rank : 89) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
ANR22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.059017 (rank : 75) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VYY1, Q8WU06 | Gene names | ANKRD22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 22. | |||||
|
ANR49_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.050273 (rank : 97) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WVL7, Q8NDF2, Q96JE5, Q9NXK7 | Gene names | ANKRD49, FGIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 49 (Fetal globin-inducing factor). | |||||
|
ANR49_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.052746 (rank : 87) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VE42, Q9Z2S1 | Gene names | Ankrd49, Fgif, Gbif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 49 (Fetal globin-inducing factor) (Fetal globin-increasing factor). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.067341 (rank : 66) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.063134 (rank : 70) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.062739 (rank : 71) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.066822 (rank : 67) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CLPB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.050178 (rank : 98) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60649 | Gene names | Clpb, Skd3 | |||
|
Domain Architecture |

|
|||||
| Description | Caseinolytic peptidase B protein homolog (Suppressor of potassium transport defect 3). | |||||
|
DPF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.067841 (rank : 65) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
DPF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.059284 (rank : 74) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
INVS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.051915 (rank : 91) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
K1333_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.058424 (rank : 78) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
K1333_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.059344 (rank : 73) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.050115 (rank : 99) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.051561 (rank : 92) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.057217 (rank : 79) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.054074 (rank : 83) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PECI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.051378 (rank : 94) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
PECI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.051191 (rank : 96) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.055965 (rank : 81) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.056195 (rank : 80) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.051946 (rank : 90) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.076788 (rank : 56) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.074933 (rank : 57) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.103365 (rank : 48) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
PHF12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.054356 (rank : 82) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.053771 (rank : 84) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.079964 (rank : 54) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
PHF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.077725 (rank : 55) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
PHF7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.068234 (rank : 64) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
PHF7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.062694 (rank : 72) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.052459 (rank : 88) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.072382 (rank : 59) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RAI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.080876 (rank : 53) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
REQU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.058916 (rank : 76) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.058469 (rank : 77) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
RFXK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.052924 (rank : 86) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14593, O95839 | Gene names | RFXANK, ANKRA1, RFXB | |||
|
Domain Architecture |
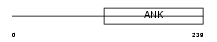
|
|||||
| Description | DNA-binding protein RFXANK (Regulatory factor X subunit B) (RFX-B) (Ankyrin repeat family A protein 1). | |||||
|
RFXK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.051446 (rank : 93) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z205 | Gene names | Rfxank, Rfxb, Tvl1 | |||
|
Domain Architecture |
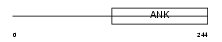
|
|||||
| Description | DNA-binding protein RFXANK (Regulatory factor X subunit B) (RFX-B) (Regulatory factor X-associated ankyrin-containing protein) (Ankyrin repeat-containing adapter protein Tvl-1). | |||||
|
SETD7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.094459 (rank : 50) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WTS6, Q9C0E6 | Gene names | SETD7, KIAA1717, SET7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7) (Set9) (SET7/9). | |||||
|
SETD7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.095243 (rank : 49) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VHL1 | Gene names | Setd7, Set7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.073674 (rank : 58) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.069034 (rank : 63) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
UHRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.050110 (rank : 100) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
SUV92_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV92_MOUSE
|
||||||
| NC score | 0.998116 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV91_HUMAN
|
||||||
| NC score | 0.986825 (rank : 3) | θ value | 9.24197e-138 (rank : 4) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
SUV91_MOUSE
|
||||||
| NC score | 0.986798 (rank : 4) | θ value | 4.14853e-138 (rank : 3) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
EZH2_HUMAN
|
||||||
| NC score | 0.691368 (rank : 5) | θ value | 3.88503e-19 (rank : 13) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15910, Q15755, Q92857 | Gene names | EZH2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
EZH1_HUMAN
|
||||||
| NC score | 0.691193 (rank : 6) | θ value | 7.82807e-20 (rank : 11) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH1_MOUSE
|
||||||
| NC score | 0.690049 (rank : 7) | θ value | 3.88503e-19 (rank : 12) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70351, Q9R089 | Gene names | Ezh1, Enx2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH2_MOUSE
|
||||||
| NC score | 0.686122 (rank : 8) | θ value | 1.92812e-18 (rank : 14) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61188, Q9R090 | Gene names | Ezh2, Enx1h | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.681190 (rank : 9) | θ value | 2.87452e-22 (rank : 10) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SETB2_HUMAN
|
||||||
| NC score | 0.636978 (rank : 10) | θ value | 8.67504e-11 (rank : 23) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
SETB1_MOUSE
|
||||||
| NC score | 0.627124 (rank : 11) | θ value | 3.40345e-15 (rank : 18) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SETB1_HUMAN
|
||||||
| NC score | 0.625135 (rank : 12) | θ value | 3.40345e-15 (rank : 17) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15047, Q96GM9 | Gene names | SETDB1, KIAA0067 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SETD8_HUMAN
|
||||||
| NC score | 0.607985 (rank : 13) | θ value | 5.62301e-10 (rank : 24) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
SETD8_MOUSE
|
||||||
| NC score | 0.607620 (rank : 14) | θ value | 9.59137e-10 (rank : 25) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q2YDW7, Q8C0J9 | Gene names | SETD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.591450 (rank : 15) | θ value | 8.93572e-24 (rank : 8) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.589334 (rank : 16) | θ value | 8.93572e-24 (rank : 9) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.572959 (rank : 17) | θ value | 4.16044e-13 (rank : 21) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.539451 (rank : 18) | θ value | 7.32683e-18 (rank : 15) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.520446 (rank : 19) | θ value | 3.07829e-16 (rank : 16) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
CBX3_MOUSE
|
||||||
| NC score | 0.422798 (rank : 20) | θ value | 6.21693e-09 (rank : 26) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CBX3_HUMAN
|
||||||
| NC score | 0.422365 (rank : 21) | θ value | 8.11959e-09 (rank : 27) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX1_HUMAN
|
||||||
| NC score | 0.420643 (rank : 22) | θ value | 5.26297e-08 (rank : 30) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| NC score | 0.420643 (rank : 23) | θ value | 5.26297e-08 (rank : 31) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | 0.418616 (rank : 24) | θ value | 1.08726e-37 (rank : 5) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.415044 (rank : 25) | θ value | 1.42001e-37 (rank : 6) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
CBX5_HUMAN
|
||||||
| NC score | 0.410837 (rank : 26) | θ value | 1.38499e-08 (rank : 28) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX5_MOUSE
|
||||||
| NC score | 0.409976 (rank : 27) | θ value | 3.08544e-08 (rank : 29) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
EHMT1_HUMAN
|
||||||
| NC score | 0.408810 (rank : 28) | θ value | 3.4976e-36 (rank : 7) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.401426 (rank : 29) | θ value | 2.43908e-13 (rank : 20) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.398949 (rank : 30) | θ value | 6.41864e-14 (rank : 19) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.381437 (rank : 31) | θ value | 9.26847e-13 (rank : 22) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
CBX7_MOUSE
|
||||||
| NC score | 0.355095 (rank : 32) | θ value | 0.000461057 (rank : 37) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX7_HUMAN
|
||||||
| NC score | 0.338432 (rank : 33) | θ value | 0.00035302 (rank : 36) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX2_MOUSE
|
||||||
| NC score | 0.327677 (rank : 34) | θ value | 0.000121331 (rank : 35) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
CBX4_HUMAN
|
||||||
| NC score | 0.314991 (rank : 35) | θ value | 9.29e-05 (rank : 32) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CBX4_MOUSE
|
||||||
| NC score | 0.312888 (rank : 36) | θ value | 9.29e-05 (rank : 33) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CBX8_HUMAN
|
||||||
| NC score | 0.309653 (rank : 37) | θ value | 0.00175202 (rank : 38) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.308278 (rank : 38) | θ value | 0.000121331 (rank : 34) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CBX8_MOUSE
|
||||||
| NC score | 0.305555 (rank : 39) | θ value | 0.00175202 (rank : 39) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QXV1 | Gene names | Cbx8, Pc3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (mPc3). | |||||
|
CBX6_HUMAN
|
||||||
| NC score | 0.299620 (rank : 40) | θ value | 0.0148317 (rank : 44) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX6_MOUSE
|
||||||
| NC score | 0.299209 (rank : 41) | θ value | 0.0148317 (rank : 45) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CDYL2_HUMAN
|
||||||
| NC score | 0.218735 (rank : 42) | θ value | 0.00509761 (rank : 42) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
CDY1_HUMAN
|
||||||
| NC score | 0.205995 (rank : 43) | θ value | 0.00228821 (rank : 40) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
CDY2_HUMAN
|
||||||
| NC score | 0.205228 (rank : 44) | θ value | 0.00509761 (rank : 41) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
CDYL1_HUMAN
|
||||||
| NC score | 0.203173 (rank : 45) | θ value | 0.125558 (rank : 48) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL_MOUSE
|
||||||
| NC score | 0.201739 (rank : 46) | θ value | 0.0431538 (rank : 47) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.168535 (rank : 47) | θ value | 0.00869519 (rank : 43) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
PHF11_HUMAN
|
||||||
| NC score | 0.103365 (rank : 48) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
SETD7_MOUSE
|
||||||
| NC score | 0.095243 (rank : 49) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VHL1 | Gene names | Setd7, Set7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7). | |||||
|
SETD7_HUMAN
|
||||||
| NC score | 0.094459 (rank : 50) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WTS6, Q9C0E6 | Gene names | SETD7, KIAA1717, SET7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7) (Set9) (SET7/9). | |||||
|
SMYD3_HUMAN
|
||||||
| NC score | 0.092520 (rank : 51) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H7B4, Q86TL8, Q8N5Z6, Q96AI5 | Gene names | SMYD3, ZMYND1, ZNFN3A1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
SMYD3_MOUSE
|
||||||
| NC score | 0.091999 (rank : 52) | θ value | 0.279714 (rank : 51) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CWR2, Q6P7V6, Q8BG90 | Gene names | Smyd3, Zmynd1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.080876 (rank : 53) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
PHF6_HUMAN
|
||||||
| NC score | 0.079964 (rank : 54) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
PHF6_MOUSE
|
||||||
| NC score | 0.077725 (rank : 55) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.076788 (rank : 56) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.074933 (rank : 57) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.073674 (rank : 58) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.072382 (rank : 59) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.069871 (rank : 60) | θ value | 0.163984 (rank : 49) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.069665 (rank : 61) | θ value | 0.163984 (rank : 50) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.069522 (rank : 62) | θ value | 0.0252991 (rank : 46) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.069034 (rank : 63) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
PHF7_HUMAN
|
||||||
| NC score | 0.068234 (rank : 64) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
DPF1_HUMAN
|
||||||
| NC score | 0.067841 (rank : 65) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.067341 (rank : 66) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.066822 (rank : 67) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.064312 (rank : 68) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.064300 (rank : 69) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.063134 (rank : 70) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.062739 (rank : 71) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
PHF7_MOUSE
|
||||||
| NC score | 0.062694 (rank : 72) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
K1333_MOUSE
|
||||||
| NC score | 0.059344 (rank : 73) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
DPF1_MOUSE
|
||||||
| NC score | 0.059284 (rank : 74) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
ANR22_HUMAN
|
||||||
| NC score | 0.059017 (rank : 75) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VYY1, Q8WU06 | Gene names | ANKRD22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 22. | |||||
|
REQU_HUMAN
|
||||||
| NC score | 0.058916 (rank : 76) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| NC score | 0.058469 (rank : 77) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
K1333_HUMAN
|
||||||
| NC score | 0.058424 (rank : 78) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.057217 (rank : 79) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.056195 (rank : 80) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.055965 (rank : 81) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.054356 (rank : 82) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.054074 (rank : 83) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.053771 (rank : 84) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
ANKY2_HUMAN
|
||||||
| NC score | 0.053153 (rank : 85) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IV38, Q96BL3 | Gene names | ANKMY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
RFXK_HUMAN
|
||||||
| NC score | 0.052924 (rank : 86) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14593, O95839 | Gene names | RFXANK, ANKRA1, RFXB | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFXANK (Regulatory factor X subunit B) (RFX-B) (Ankyrin repeat family A protein 1). | |||||
|
ANR49_MOUSE
|
||||||
| NC score | 0.052746 (rank : 87) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VE42, Q9Z2S1 | Gene names | Ankrd49, Fgif, Gbif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 49 (Fetal globin-inducing factor) (Fetal globin-increasing factor). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.052459 (rank : 88) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
ANKY2_MOUSE
|
||||||
| NC score | 0.052270 (rank : 89) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.051946 (rank : 90) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.051915 (rank : 91) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.051561 (rank : 92) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
RFXK_MOUSE
|
||||||
| NC score | 0.051446 (rank : 93) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z205 | Gene names | Rfxank, Rfxb, Tvl1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFXANK (Regulatory factor X subunit B) (RFX-B) (Regulatory factor X-associated ankyrin-containing protein) (Ankyrin repeat-containing adapter protein Tvl-1). | |||||
|
PECI_HUMAN
|
||||||
| NC score | 0.051378 (rank : 94) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
ANKR6_HUMAN
|
||||||
| NC score | 0.051330 (rank : 95) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
PECI_MOUSE
|
||||||
| NC score | 0.051191 (rank : 96) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
ANR49_HUMAN
|
||||||
| NC score | 0.050273 (rank : 97) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WVL7, Q8NDF2, Q96JE5, Q9NXK7 | Gene names | ANKRD49, FGIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 49 (Fetal globin-inducing factor). | |||||
|
CLPB_MOUSE
|
||||||
| NC score | 0.050178 (rank : 98) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60649 | Gene names | Clpb, Skd3 | |||
|
Domain Architecture |

|
|||||
| Description | Caseinolytic peptidase B protein homolog (Suppressor of potassium transport defect 3). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.050115 (rank : 99) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
UHRF1_MOUSE
|
||||||
| NC score | 0.050110 (rank : 100) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
IFNA4_MOUSE
|
||||||
| NC score | 0.010379 (rank : 101) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07351 | Gene names | Ifna4 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon alpha-4 precursor. | |||||
|
SO1B3_HUMAN
|
||||||
| NC score | 0.005346 (rank : 102) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPD5 | Gene names | SLCO1B3, LST2, SLC21A8 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 1B3 (Solute carrier family 21 member 8) (Organic anion transporter 8) (Organic anion-transporting polypeptide 8) (OATP8) (Liver-specific organic anion transporter 2) (LST-2). | |||||