Please be patient as the page loads
|
CBX2_MOUSE
|
||||||
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CBX2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.928138 (rank : 2) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CBX2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
CBX4_HUMAN
|
||||||
| θ value | 3.07116e-24 (rank : 3) | NC score | 0.804034 (rank : 8) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CBX4_MOUSE
|
||||||
| θ value | 8.93572e-24 (rank : 4) | NC score | 0.801622 (rank : 9) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CBX7_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 5) | NC score | 0.849722 (rank : 4) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX8_HUMAN
|
||||||
| θ value | 3.17815e-21 (rank : 6) | NC score | 0.810413 (rank : 6) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
CBX8_MOUSE
|
||||||
| θ value | 3.17815e-21 (rank : 7) | NC score | 0.811730 (rank : 5) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXV1 | Gene names | Cbx8, Pc3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (mPc3). | |||||
|
CBX6_HUMAN
|
||||||
| θ value | 5.42112e-21 (rank : 8) | NC score | 0.798726 (rank : 10) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX6_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 9) | NC score | 0.807575 (rank : 7) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX7_MOUSE
|
||||||
| θ value | 1.74391e-19 (rank : 10) | NC score | 0.862824 (rank : 3) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX5_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 11) | NC score | 0.675740 (rank : 11) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX5_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 12) | NC score | 0.671188 (rank : 14) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX1_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 13) | NC score | 0.672389 (rank : 12) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 14) | NC score | 0.672389 (rank : 13) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CDYL1_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 15) | NC score | 0.413664 (rank : 17) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CBX3_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 16) | NC score | 0.643400 (rank : 15) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX3_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 17) | NC score | 0.631983 (rank : 16) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 18) | NC score | 0.238584 (rank : 26) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
CDYL2_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 19) | NC score | 0.408113 (rank : 18) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
SUV91_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 20) | NC score | 0.331364 (rank : 23) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV92_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 21) | NC score | 0.345521 (rank : 22) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
CDYL_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 22) | NC score | 0.403814 (rank : 19) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 23) | NC score | 0.178296 (rank : 27) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CDY1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 24) | NC score | 0.383328 (rank : 21) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
CDY2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 25) | NC score | 0.384039 (rank : 20) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 26) | NC score | 0.170225 (rank : 29) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
SUV92_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 27) | NC score | 0.327677 (rank : 24) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 28) | NC score | 0.176001 (rank : 28) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
SUV91_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 29) | NC score | 0.300418 (rank : 25) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
CDCA7_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 30) | NC score | 0.086139 (rank : 33) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BWT1, Q53EW5, Q580W9, Q658K4, Q658N4, Q8NBY9, Q96BV8, Q96SP5 | Gene names | CDCA7, JPO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle-associated protein 7 (Protein JPO1). | |||||
|
MS3L1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 31) | NC score | 0.133323 (rank : 30) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.052252 (rank : 53) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
ANKS6_MOUSE
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.026721 (rank : 75) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
RANB3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 34) | NC score | 0.047370 (rank : 56) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CT10, Q3TIS4, Q3U2G4, Q3UYG7 | Gene names | Ranbp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
K2027_MOUSE
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.047088 (rank : 57) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BWS5, Q69Z30 | Gene names | Kiaa2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 36) | NC score | 0.019387 (rank : 88) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.043246 (rank : 59) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.044950 (rank : 58) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
DCAK3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.004987 (rank : 122) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BWQ5, Q1A748 | Gene names | Dcamkl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL3 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 3) (CLICK-I and II-related) (CLr). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.015057 (rank : 99) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.039143 (rank : 61) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.048752 (rank : 55) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
M3K12_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.002366 (rank : 130) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.034049 (rank : 69) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.036087 (rank : 66) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
DNL3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.021164 (rank : 84) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
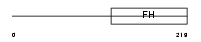
FOXD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.007325 (rank : 116) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60548 | Gene names | FOXD2, FKHL17, FREAC9 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein D2 (Forkhead-related protein FKHL17) (Forkhead- related transcription factor 9) (FREAC-9). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.032848 (rank : 70) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.029643 (rank : 73) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.048951 (rank : 54) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.022139 (rank : 83) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.039233 (rank : 60) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
EP400_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.062894 (rank : 42) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.034472 (rank : 68) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.019444 (rank : 87) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
MYLK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.004400 (rank : 123) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15746, O95796, O95797, O95798, O95799, Q14844, Q16794, Q5MY99, Q5MYA0, Q7Z4J0, Q9C0L5, Q9UBG5, Q9UIT9 | Gene names | MYLK, MLCK | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
NCA11_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.014091 (rank : 103) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.037480 (rank : 63) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
VPS72_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.036226 (rank : 65) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
VPS72_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.035643 (rank : 67) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.007697 (rank : 115) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.024578 (rank : 80) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.037161 (rank : 64) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PAF49_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.024899 (rank : 78) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q76KJ5, Q8K0Y9 | Gene names | Cd3eap, Ase1, Paf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.023298 (rank : 82) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
POT15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.018565 (rank : 92) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
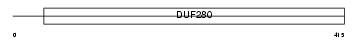
ACM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.002122 (rank : 131) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 917 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12657, Q8BJN3 | Gene names | Chrm1, Chrm-1 | |||
|
Domain Architecture |
|
|||||
| Description | Muscarinic acetylcholine receptor M1. | |||||
|
ANR41_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.014553 (rank : 102) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
CD2L5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.002858 (rank : 127) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.015295 (rank : 97) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CSKP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.004195 (rank : 125) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1022 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14936, O43215, Q9NYB3 | Gene names | CASK, LIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (hCASK) (Calcium/calmodulin-dependent serine protein kinase) (Lin-2 homolog). | |||||
|
CSKP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.004213 (rank : 124) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O70589, O70588 | Gene names | Cask | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (Calcium/calmodulin-dependent serine protein kinase). | |||||
|
GRIN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.030318 (rank : 72) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60269, Q5SVF0 | Gene names | GPRIN2, KIAA0514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 2 (GRIN2). | |||||
|
LIPA3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.007170 (rank : 117) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
PDE1C_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.009644 (rank : 107) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
RBM12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.016885 (rank : 95) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.057888 (rank : 46) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
DCAK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.003269 (rank : 126) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C098 | Gene names | DCAMKL3, KIAA1765 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL3 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 3). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.009428 (rank : 108) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.019807 (rank : 86) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.005556 (rank : 120) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.017439 (rank : 93) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NR4A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.005629 (rank : 119) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22736 | Gene names | NR4A1, GFRP1, HMR, NAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A1 (Orphan nuclear receptor HMR) (Early response protein NAK1) (TR3 orphan receptor) (ST-59). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.038687 (rank : 62) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RFTN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.014845 (rank : 101) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14699, Q496Y2, Q4QQI7, Q5JB48, Q7Z7P2 | Gene names | RFTN1, KIAA0084 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin (Raft-linking protein) (Cell migration-inducing gene 2 protein). | |||||
|
SENP6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.018697 (rank : 91) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9GZR1, O94891, Q8TBY4, Q9UJV5 | Gene names | SENP6, KIAA0797, SSP1, SUSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 6 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP6) (SUMO-1-specific protease 1). | |||||
|
SP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | -0.001169 (rank : 134) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02447, Q8TD56, Q9BQR1 | Gene names | SP3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp3 (SPR-2). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.024622 (rank : 79) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
CABL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.012044 (rank : 104) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.025381 (rank : 77) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
DLGP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.008937 (rank : 112) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95886, Q5TDD5, Q9H3X7 | Gene names | DLGAP3, DAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.006570 (rank : 118) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
KLF16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | -0.000026 (rank : 133) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BXK1 | Gene names | KLF16, BTEB4, NSLP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 16 (Transcription factor BTEB4) (Basic transcription element-binding protein 4) (BTE-binding protein 4) (Novel Sp1-like zinc finger transcription factor 2) (Transcription factor NSLP2). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.026653 (rank : 76) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NR4A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.005169 (rank : 121) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P12813 | Gene names | Nr4a1, Gfrp, Hmr, N10, Nur77 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A1 (Orphan nuclear receptor HMR) (Nuclear hormone receptor NUR/77) (N10 nuclear protein). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.014900 (rank : 100) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.009223 (rank : 109) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.001661 (rank : 132) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.028159 (rank : 74) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.011097 (rank : 105) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.018829 (rank : 89) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
VGLL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.017391 (rank : 94) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99NC0 | Gene names | Vgll1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor vestigial-like protein 1 (Vgl-1) (Vestigial- related factor). | |||||
|
ZN185_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.016038 (rank : 96) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15231, O00345, Q9NSD2 | Gene names | ZNF185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein ZNF185) (P1-A). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.008488 (rank : 114) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.024143 (rank : 81) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
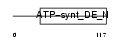
ATPD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.015129 (rank : 98) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30049 | Gene names | ATP5D | |||
|
Domain Architecture |
|
|||||
| Description | ATP synthase delta chain, mitochondrial precursor (EC 3.6.3.14). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.083503 (rank : 34) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.073018 (rank : 40) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.073635 (rank : 39) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
DUX4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.002654 (rank : 129) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBX2 | Gene names | DUX4, DUX10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double homeobox protein 4 (Double homeobox protein 10) (Double homeobox protein 4/10). | |||||
|
H2B3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.008691 (rank : 113) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D2U9 | Gene names | Hist3h2ba | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2B type 3-A. | |||||
|
H2B3B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.009110 (rank : 110) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N257, Q3ZCP6, Q5TA30 | Gene names | HIST3H2BB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2B type 3-B (H2B type 12). | |||||
|
H2B3B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.009110 (rank : 111) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGP0, Q3B7Z7 | Gene names | Hist3h2bb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2B type 3-B. | |||||
|
HSH2D_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.009897 (rank : 106) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96JZ2, Q6ZNG7 | Gene names | HSH2D, ALX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
ICAL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.018740 (rank : 90) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
IF35_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.032540 (rank : 71) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
KIF2C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.002681 (rank : 128) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99661, Q96C18, Q96HB8, Q9BWV8 | Gene names | KIF2C, KNSL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF2C (Mitotic centromere-associated kinesin) (MCAK) (Kinesin-like protein 6). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.021010 (rank : 85) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.055215 (rank : 50) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
AUHM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.060557 (rank : 44) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLZ3, Q80YD7, Q8QZS0, Q9CY78, Q9D155 | Gene names | Auh | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding enoyl-CoA hydratase) (muAUH). | |||||
|
AUMH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.059820 (rank : 45) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13825, Q8WUE4 | Gene names | AUH | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding protein/enoyl-CoA hydratase). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.067638 (rank : 41) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.062122 (rank : 43) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.057267 (rank : 47) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.056586 (rank : 48) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.056311 (rank : 49) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
ECH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.076089 (rank : 36) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13011, Q8WVX0, Q96EZ9 | Gene names | ECH1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.078463 (rank : 35) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35459, Q5M8P6 | Gene names | Ech1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECHM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.074006 (rank : 38) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECHM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.074399 (rank : 37) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BH95, Q3TJK2, Q6GQS2, Q6PEN1, Q99LX7 | Gene names | Echs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
EP400_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.054438 (rank : 51) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
PECI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.113846 (rank : 31) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
PECI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.113121 (rank : 32) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.054376 (rank : 52) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
CBX2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.928138 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CBX7_MOUSE
|
||||||
| NC score | 0.862824 (rank : 3) | θ value | 1.74391e-19 (rank : 10) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX7_HUMAN
|
||||||
| NC score | 0.849722 (rank : 4) | θ value | 1.42661e-21 (rank : 5) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX8_MOUSE
|
||||||
| NC score | 0.811730 (rank : 5) | θ value | 3.17815e-21 (rank : 7) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXV1 | Gene names | Cbx8, Pc3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (mPc3). | |||||
|
CBX8_HUMAN
|
||||||
| NC score | 0.810413 (rank : 6) | θ value | 3.17815e-21 (rank : 6) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
CBX6_MOUSE
|
||||||
| NC score | 0.807575 (rank : 7) | θ value | 5.42112e-21 (rank : 9) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX4_HUMAN
|
||||||
| NC score | 0.804034 (rank : 8) | θ value | 3.07116e-24 (rank : 3) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CBX4_MOUSE
|
||||||
| NC score | 0.801622 (rank : 9) | θ value | 8.93572e-24 (rank : 4) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CBX6_HUMAN
|
||||||
| NC score | 0.798726 (rank : 10) | θ value | 5.42112e-21 (rank : 8) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX5_MOUSE
|
||||||
| NC score | 0.675740 (rank : 11) | θ value | 1.2105e-12 (rank : 11) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX1_HUMAN
|
||||||
| NC score | 0.672389 (rank : 12) | θ value | 3.29651e-10 (rank : 13) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| NC score | 0.672389 (rank : 13) | θ value | 3.29651e-10 (rank : 14) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX5_HUMAN
|
||||||
| NC score | 0.671188 (rank : 14) | θ value | 1.02475e-11 (rank : 12) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX3_HUMAN
|
||||||
| NC score | 0.643400 (rank : 15) | θ value | 8.97725e-08 (rank : 16) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX3_MOUSE
|
||||||
| NC score | 0.631983 (rank : 16) | θ value | 4.45536e-07 (rank : 17) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CDYL1_HUMAN
|
||||||
| NC score | 0.413664 (rank : 17) | θ value | 6.87365e-08 (rank : 15) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL2_HUMAN
|
||||||
| NC score | 0.408113 (rank : 18) | θ value | 8.40245e-06 (rank : 19) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
CDYL_MOUSE
|
||||||
| NC score | 0.403814 (rank : 19) | θ value | 5.44631e-05 (rank : 22) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDY2_HUMAN
|
||||||
| NC score | 0.384039 (rank : 20) | θ value | 0.000121331 (rank : 25) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
CDY1_HUMAN
|
||||||
| NC score | 0.383328 (rank : 21) | θ value | 0.000121331 (rank : 24) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
SUV92_MOUSE
|
||||||
| NC score | 0.345521 (rank : 22) | θ value | 2.44474e-05 (rank : 21) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV91_MOUSE
|
||||||
| NC score | 0.331364 (rank : 23) | θ value | 1.43324e-05 (rank : 20) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV92_HUMAN
|
||||||
| NC score | 0.327677 (rank : 24) | θ value | 0.000121331 (rank : 27) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV91_HUMAN
|
||||||
| NC score | 0.300418 (rank : 25) | θ value | 0.00298849 (rank : 29) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.238584 (rank : 26) | θ value | 5.81887e-07 (rank : 18) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.178296 (rank : 27) | θ value | 5.44631e-05 (rank : 23) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.176001 (rank : 28) | θ value | 0.000158464 (rank : 28) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.170225 (rank : 29) | θ value | 0.000121331 (rank : 26) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
MS3L1_MOUSE
|
||||||
| NC score | 0.133323 (rank : 30) | θ value | 0.0563607 (rank : 31) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
PECI_HUMAN
|
||||||
| NC score | 0.113846 (rank : 31) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
PECI_MOUSE
|
||||||
| NC score | 0.113121 (rank : 32) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
CDCA7_HUMAN
|
||||||
| NC score | 0.086139 (rank : 33) | θ value | 0.0252991 (rank : 30) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BWT1, Q53EW5, Q580W9, Q658K4, Q658N4, Q8NBY9, Q96BV8, Q96SP5 | Gene names | CDCA7, JPO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle-associated protein 7 (Protein JPO1). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.083503 (rank : 34) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
ECH1_MOUSE
|
||||||
| NC score | 0.078463 (rank : 35) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35459, Q5M8P6 | Gene names | Ech1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECH1_HUMAN
|
||||||
| NC score | 0.076089 (rank : 36) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13011, Q8WVX0, Q96EZ9 | Gene names | ECH1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECHM_MOUSE
|
||||||
| NC score | 0.074399 (rank : 37) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BH95, Q3TJK2, Q6GQS2, Q6PEN1, Q99LX7 | Gene names | Echs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECHM_HUMAN
|
||||||
| NC score | 0.074006 (rank : 38) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.073635 (rank : 39) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.073018 (rank : 40) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.067638 (rank : 41) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.062894 (rank : 42) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.062122 (rank : 43) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
AUHM_MOUSE
|
||||||
| NC score | 0.060557 (rank : 44) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLZ3, Q80YD7, Q8QZS0, Q9CY78, Q9D155 | Gene names | Auh | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding enoyl-CoA hydratase) (muAUH). | |||||
|
AUMH_HUMAN
|
||||||
| NC score | 0.059820 (rank : 45) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13825, Q8WUE4 | Gene names | AUH | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding protein/enoyl-CoA hydratase). | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.057888 (rank : 46) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.057267 (rank : 47) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.056586 (rank : 48) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.056311 (rank : 49) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.055215 (rank : 50) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.054438 (rank : 51) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.054376 (rank : 52) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.052252 (rank : 53) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.048951 (rank : 54) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.048752 (rank : 55) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
RANB3_MOUSE
|
||||||
| NC score | 0.047370 (rank : 56) | θ value | 0.21417 (rank : 34) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CT10, Q3TIS4, Q3U2G4, Q3UYG7 | Gene names | Ranbp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
K2027_MOUSE
|
||||||
| NC score | 0.047088 (rank : 57) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BWS5, Q69Z30 | Gene names | Kiaa2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.044950 (rank : 58) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.043246 (rank : 59) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.039233 (rank : 60) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.039143 (rank : 61) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.038687 (rank : 62) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.037480 (rank : 63) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.037161 (rank : 64) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
VPS72_HUMAN
|
||||||
| NC score | 0.036226 (rank : 65) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.036087 (rank : 66) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
VPS72_MOUSE
|
||||||
| NC score | 0.035643 (rank : 67) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.034472 (rank : 68) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.034049 (rank : 69) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.032848 (rank : 70) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
IF35_MOUSE
|
||||||
| NC score | 0.032540 (rank : 71) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
GRIN2_HUMAN
|
||||||
| NC score | 0.030318 (rank : 72) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60269, Q5SVF0 | Gene names | GPRIN2, KIAA0514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 2 (GRIN2). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.029643 (rank : 73) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.028159 (rank : 74) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
ANKS6_MOUSE
|
||||||
| NC score | 0.026721 (rank : 75) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.026653 (rank : 76) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.025381 (rank : 77) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
PAF49_MOUSE
|
||||||
| NC score | 0.024899 (rank : 78) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q76KJ5, Q8K0Y9 | Gene names | Cd3eap, Ase1, Paf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.024622 (rank : 79) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.024578 (rank : 80) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.024143 (rank : 81) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.023298 (rank : 82) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.022139 (rank : 83) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
DNL3_HUMAN
|
||||||
| NC score | 0.021164 (rank : 84) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.021010 (rank : 85) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.019807 (rank : 86) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.019444 (rank : 87) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.019387 (rank : 88) | θ value | 0.279714 (rank : 36) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.018829 (rank : 89) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
ICAL_HUMAN
|
||||||
| NC score | 0.018740 (rank : 90) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
SENP6_HUMAN
|
||||||
| NC score | 0.018697 (rank : 91) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9GZR1, O94891, Q8TBY4, Q9UJV5 | Gene names | SENP6, KIAA0797, SSP1, SUSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 6 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP6) (SUMO-1-specific protease 1). | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.018565 (rank : 92) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.017439 (rank : 93) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
VGLL1_MOUSE
|
||||||
| NC score | 0.017391 (rank : 94) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99NC0 | Gene names | Vgll1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor vestigial-like protein 1 (Vgl-1) (Vestigial- related factor). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.016885 (rank : 95) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
ZN185_HUMAN
|
||||||
| NC score | 0.016038 (rank : 96) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15231, O00345, Q9NSD2 | Gene names | ZNF185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein ZNF185) (P1-A). | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.015295 (rank : 97) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
ATPD_HUMAN
|
||||||
| NC score | 0.015129 (rank : 98) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30049 | Gene names | ATP5D | |||
|
Domain Architecture |

|
|||||
| Description | ATP synthase delta chain, mitochondrial precursor (EC 3.6.3.14). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.015057 (rank : 99) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.014900 (rank : 100) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RFTN1_HUMAN
|
||||||
| NC score | 0.014845 (rank : 101) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14699, Q496Y2, Q4QQI7, Q5JB48, Q7Z7P2 | Gene names | RFTN1, KIAA0084 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin (Raft-linking protein) (Cell migration-inducing gene 2 protein). | |||||
|
ANR41_HUMAN
|
||||||
| NC score | 0.014553 (rank : 102) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
NCA11_MOUSE
|
||||||
| NC score | 0.014091 (rank : 103) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
CABL2_HUMAN
|
||||||
| NC score | 0.012044 (rank : 104) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.011097 (rank : 105) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
HSH2D_HUMAN
|
||||||
| NC score | 0.009897 (rank : 106) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96JZ2, Q6ZNG7 | Gene names | HSH2D, ALX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
PDE1C_MOUSE
|
||||||
| NC score | 0.009644 (rank : 107) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.009428 (rank : 108) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.009223 (rank : 109) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
H2B3B_HUMAN
|
||||||
| NC score | 0.009110 (rank : 110) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N257, Q3ZCP6, Q5TA30 | Gene names | HIST3H2BB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2B type 3-B (H2B type 12). | |||||
|
H2B3B_MOUSE
|
||||||
| NC score | 0.009110 (rank : 111) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGP0, Q3B7Z7 | Gene names | Hist3h2bb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2B type 3-B. | |||||
|
DLGP3_HUMAN
|
||||||
| NC score | 0.008937 (rank : 112) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95886, Q5TDD5, Q9H3X7 | Gene names | DLGAP3, DAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
H2B3A_MOUSE
|
||||||
| NC score | 0.008691 (rank : 113) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D2U9 | Gene names | Hist3h2ba | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone H2B type 3-A. | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.008488 (rank : 114) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.007697 (rank : 115) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
FOXD2_HUMAN
|
||||||
| NC score | 0.007325 (rank : 116) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60548 | Gene names | FOXD2, FKHL17, FREAC9 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D2 (Forkhead-related protein FKHL17) (Forkhead- related transcription factor 9) (FREAC-9). | |||||
|
LIPA3_MOUSE
|
||||||
| NC score | 0.007170 (rank : 117) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.006570 (rank : 118) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
NR4A1_HUMAN
|
||||||
| NC score | 0.005629 (rank : 119) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22736 | Gene names | NR4A1, GFRP1, HMR, NAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A1 (Orphan nuclear receptor HMR) (Early response protein NAK1) (TR3 orphan receptor) (ST-59). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.005556 (rank : 120) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
NR4A1_MOUSE
|
||||||
| NC score | 0.005169 (rank : 121) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P12813 | Gene names | Nr4a1, Gfrp, Hmr, N10, Nur77 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A1 (Orphan nuclear receptor HMR) (Nuclear hormone receptor NUR/77) (N10 nuclear protein). | |||||
|
DCAK3_MOUSE
|
||||||
| NC score | 0.004987 (rank : 122) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BWQ5, Q1A748 | Gene names | Dcamkl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL3 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 3) (CLICK-I and II-related) (CLr). | |||||
|
MYLK_HUMAN
|
||||||
| NC score | 0.004400 (rank : 123) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15746, O95796, O95797, O95798, O95799, Q14844, Q16794, Q5MY99, Q5MYA0, Q7Z4J0, Q9C0L5, Q9UBG5, Q9UIT9 | Gene names | MYLK, MLCK | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
CSKP_MOUSE
|
||||||
| NC score | 0.004213 (rank : 124) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O70589, O70588 | Gene names | Cask | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (Calcium/calmodulin-dependent serine protein kinase). | |||||
|
CSKP_HUMAN
|
||||||
| NC score | 0.004195 (rank : 125) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1022 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14936, O43215, Q9NYB3 | Gene names | CASK, LIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (hCASK) (Calcium/calmodulin-dependent serine protein kinase) (Lin-2 homolog). | |||||
|
DCAK3_HUMAN
|
||||||
| NC score | 0.003269 (rank : 126) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C098 | Gene names | DCAMKL3, KIAA1765 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL3 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 3). | |||||
|
CD2L5_MOUSE
|
||||||
| NC score | 0.002858 (rank : 127) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
KIF2C_HUMAN
|
||||||
| NC score | 0.002681 (rank : 128) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99661, Q96C18, Q96HB8, Q9BWV8 | Gene names | KIF2C, KNSL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF2C (Mitotic centromere-associated kinesin) (MCAK) (Kinesin-like protein 6). | |||||
|
DUX4_HUMAN
|
||||||
| NC score | 0.002654 (rank : 129) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBX2 | Gene names | DUX4, DUX10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double homeobox protein 4 (Double homeobox protein 10) (Double homeobox protein 4/10). | |||||
|
M3K12_MOUSE
|
||||||
| NC score | 0.002366 (rank : 130) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
ACM1_MOUSE
|
||||||
| NC score | 0.002122 (rank : 131) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 917 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12657, Q8BJN3 | Gene names | Chrm1, Chrm-1 | |||
|
Domain Architecture |

|
|||||
| Description | Muscarinic acetylcholine receptor M1. | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.001661 (rank : 132) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
KLF16_HUMAN
|
||||||
| NC score | -0.000026 (rank : 133) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BXK1 | Gene names | KLF16, BTEB4, NSLP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 16 (Transcription factor BTEB4) (Basic transcription element-binding protein 4) (BTE-binding protein 4) (Novel Sp1-like zinc finger transcription factor 2) (Transcription factor NSLP2). | |||||
|
SP3_HUMAN
|
||||||
| NC score | -0.001169 (rank : 134) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02447, Q8TD56, Q9BQR1 | Gene names | SP3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp3 (SPR-2). | |||||