Please be patient as the page loads
|
PAF49_MOUSE
|
||||||
| SwissProt Accessions | Q76KJ5, Q8K0Y9 | Gene names | Cd3eap, Ase1, Paf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PAF49_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q76KJ5, Q8K0Y9 | Gene names | Cd3eap, Ase1, Paf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein). | |||||
|
PAF49_HUMAN
|
||||||
| θ value | 4.64023e-97 (rank : 2) | NC score | 0.821925 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 3) | NC score | 0.095272 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
TMPSD_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 4) | NC score | 0.022032 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
DAAM1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 5) | NC score | 0.050054 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
JIP2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 6) | NC score | 0.085303 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 7) | NC score | 0.037029 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
PRCC_HUMAN
|
||||||
| θ value | 0.279714 (rank : 8) | NC score | 0.089536 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
FRAT1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.081037 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70339 | Gene names | Frat1 | |||
|
Domain Architecture |
|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
BARH1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.022339 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BZE3, Q9NY88 | Gene names | BARHL1 | |||
|
Domain Architecture |
|
|||||
| Description | BarH-like 1 homeobox protein. | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.074863 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.018564 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
CABL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.045380 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.021037 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
FRAT1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.076739 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92837, Q8NE74, Q8TDW9 | Gene names | FRAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
TENS4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.050137 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
SIA7A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.034401 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
ZNHI4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.059966 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PT3, Q9CY38 | Gene names | Znhit4, Hmga1l4, Papa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.017902 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
THUM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.044258 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99J36, Q3U6Y3 | Gene names | Thumpd1 | |||
|
Domain Architecture |

|
|||||
| Description | THUMP domain-containing protein 1. | |||||
|
CBX2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.024899 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.017044 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.024093 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.023909 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
XRCC4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.034244 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
AQR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.023419 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFQ3, P97871, Q3U9N1, Q3ULE8, Q80TX8 | Gene names | Aqr, Kiaa0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius. | |||||
|
NFAC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.018319 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
AL2S4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.044299 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
FRS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.019283 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WU20, O43558, Q7LDQ6 | Gene names | FRS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT). | |||||
|
GATA3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.015204 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23772 | Gene names | Gata3, Gata-3 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-acting T-cell-specific transcription factor GATA-3 (GATA-binding factor 3). | |||||
|
HXD10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.011499 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28359, Q91WU9 | Gene names | Hoxd10, Hox-4.5, Hoxd-10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D10 (Hox-4.5) (Hox-5.3). | |||||
|
JIP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.058532 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
LTB1S_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.005683 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
PARG_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.023965 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.023778 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.017556 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
CEBPB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.016397 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
FOXN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.008550 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00409, Q96II7, Q9UIE7 | Gene names | CHES1, FOXN3 | |||
|
Domain Architecture |
|
|||||
| Description | Checkpoint suppressor 1 (Forkhead box protein N3). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.015443 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.023529 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
WHRN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.018865 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.019386 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
BARH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.015395 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63157, Q9JK26 | Gene names | Barhl1, Bhx1, Mbh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BarH-like 1 homeobox protein (Bar-class homeodomain protein MBH2) (BarH-related homeobox protein 1). | |||||
|
CD2B2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.019391 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CWK3, Q3TJX5 | Gene names | Cd2bp2 | |||
|
Domain Architecture |

|
|||||
| Description | CD2 antigen cytoplasmic tail-binding protein 2 (CD2 cytoplasmic domain-binding protein) (CD2 tail-binding protein). | |||||
|
HIPK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.003702 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H422, O14632, Q2PBG4, Q2PBG5, Q92632, Q9HAS2 | Gene names | HIPK3, DYRK6, FIST3, PKY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeodomain-interacting protein kinase 3 (EC 2.7.11.1) (Homolog of protein kinase YAK1) (Fas-interacting serine/threonine-protein kinase) (FIST) (Androgen receptor-interacting nuclear protein kinase) (ANPK). | |||||
|
HIPK3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.003646 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ERH7, O88906, Q9QZR2 | Gene names | Hipk3, Fist3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeodomain-interacting protein kinase 3 (EC 2.7.11.1) (Fas- interacting serine/threonine-protein kinase) (FIST) (Nuclear body- associated kinase 3) (Nbak3) (Androgen receptor-interacting nuclear protein kinase) (ANPK). | |||||
|
HKR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.000267 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10074 | Gene names | HKR3 | |||
|
Domain Architecture |
|
|||||
| Description | Krueppel-related zinc finger protein 3 (Protein HKR3). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.020213 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
FM14A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.057954 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9H2X8, Q8TBD7, Q9NYL0 | Gene names | FAM14A, TLH29 | |||
|
Domain Architecture |
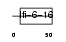
|
|||||
| Description | Protein FAM14A precursor (Protein ISG12(b)) (Protein TLH29) (pIFI27- like protein). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.051021 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PAF49_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q76KJ5, Q8K0Y9 | Gene names | Cd3eap, Ase1, Paf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein). | |||||
|
PAF49_HUMAN
|
||||||
| NC score | 0.821925 (rank : 2) | θ value | 4.64023e-97 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.095272 (rank : 3) | θ value | 0.00298849 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PRCC_HUMAN
|
||||||
| NC score | 0.089536 (rank : 4) | θ value | 0.279714 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
JIP2_MOUSE
|
||||||
| NC score | 0.085303 (rank : 5) | θ value | 0.0252991 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
FRAT1_MOUSE
|
||||||
| NC score | 0.081037 (rank : 6) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70339 | Gene names | Frat1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
FRAT1_HUMAN
|
||||||
| NC score | 0.076739 (rank : 7) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92837, Q8NE74, Q8TDW9 | Gene names | FRAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.074863 (rank : 8) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
ZNHI4_MOUSE
|
||||||
| NC score | 0.059966 (rank : 9) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PT3, Q9CY38 | Gene names | Znhit4, Hmga1l4, Papa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
JIP2_HUMAN
|
||||||
| NC score | 0.058532 (rank : 10) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
FM14A_HUMAN
|
||||||
| NC score | 0.057954 (rank : 11) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9H2X8, Q8TBD7, Q9NYL0 | Gene names | FAM14A, TLH29 | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAM14A precursor (Protein ISG12(b)) (Protein TLH29) (pIFI27- like protein). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.051021 (rank : 12) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
TENS4_MOUSE
|
||||||
| NC score | 0.050137 (rank : 13) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
DAAM1_HUMAN
|
||||||
| NC score | 0.050054 (rank : 14) | θ value | 0.0193708 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
CABL1_HUMAN
|
||||||
| NC score | 0.045380 (rank : 15) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
AL2S4_MOUSE
|
||||||
| NC score | 0.044299 (rank : 16) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
THUM1_MOUSE
|
||||||
| NC score | 0.044258 (rank : 17) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99J36, Q3U6Y3 | Gene names | Thumpd1 | |||
|
Domain Architecture |

|
|||||
| Description | THUMP domain-containing protein 1. | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.037029 (rank : 18) | θ value | 0.21417 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
SIA7A_HUMAN
|
||||||
| NC score | 0.034401 (rank : 19) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
XRCC4_HUMAN
|
||||||
| NC score | 0.034244 (rank : 20) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
CBX2_MOUSE
|
||||||
| NC score | 0.024899 (rank : 21) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.024093 (rank : 22) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
PARG_MOUSE
|
||||||
| NC score | 0.023965 (rank : 23) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.023909 (rank : 24) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.023778 (rank : 25) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.023529 (rank : 26) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
AQR_MOUSE
|
||||||
| NC score | 0.023419 (rank : 27) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFQ3, P97871, Q3U9N1, Q3ULE8, Q80TX8 | Gene names | Aqr, Kiaa0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius. | |||||
|
BARH1_HUMAN
|
||||||
| NC score | 0.022339 (rank : 28) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BZE3, Q9NY88 | Gene names | BARHL1 | |||
|
Domain Architecture |

|
|||||
| Description | BarH-like 1 homeobox protein. | |||||
|
TMPSD_MOUSE
|
||||||
| NC score | 0.022032 (rank : 29) | θ value | 0.0148317 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.021037 (rank : 30) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.020213 (rank : 31) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
CD2B2_MOUSE
|
||||||
| NC score | 0.019391 (rank : 32) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CWK3, Q3TJX5 | Gene names | Cd2bp2 | |||
|
Domain Architecture |

|
|||||
| Description | CD2 antigen cytoplasmic tail-binding protein 2 (CD2 cytoplasmic domain-binding protein) (CD2 tail-binding protein). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.019386 (rank : 33) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
FRS2_HUMAN
|
||||||
| NC score | 0.019283 (rank : 34) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WU20, O43558, Q7LDQ6 | Gene names | FRS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT). | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.018865 (rank : 35) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.018564 (rank : 36) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
NFAC4_HUMAN
|
||||||
| NC score | 0.018319 (rank : 37) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.017902 (rank : 38) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.017556 (rank : 39) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.017044 (rank : 40) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
CEBPB_HUMAN
|
||||||
| NC score | 0.016397 (rank : 41) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.015443 (rank : 42) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
BARH1_MOUSE
|
||||||
| NC score | 0.015395 (rank : 43) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63157, Q9JK26 | Gene names | Barhl1, Bhx1, Mbh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BarH-like 1 homeobox protein (Bar-class homeodomain protein MBH2) (BarH-related homeobox protein 1). | |||||
|
GATA3_MOUSE
|
||||||
| NC score | 0.015204 (rank : 44) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23772 | Gene names | Gata3, Gata-3 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-acting T-cell-specific transcription factor GATA-3 (GATA-binding factor 3). | |||||
|
HXD10_MOUSE
|
||||||
| NC score | 0.011499 (rank : 45) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28359, Q91WU9 | Gene names | Hoxd10, Hox-4.5, Hoxd-10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D10 (Hox-4.5) (Hox-5.3). | |||||
|
FOXN3_HUMAN
|
||||||
| NC score | 0.008550 (rank : 46) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00409, Q96II7, Q9UIE7 | Gene names | CHES1, FOXN3 | |||
|
Domain Architecture |

|
|||||
| Description | Checkpoint suppressor 1 (Forkhead box protein N3). | |||||
|
LTB1S_HUMAN
|
||||||
| NC score | 0.005683 (rank : 47) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
HIPK3_HUMAN
|
||||||
| NC score | 0.003702 (rank : 48) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H422, O14632, Q2PBG4, Q2PBG5, Q92632, Q9HAS2 | Gene names | HIPK3, DYRK6, FIST3, PKY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeodomain-interacting protein kinase 3 (EC 2.7.11.1) (Homolog of protein kinase YAK1) (Fas-interacting serine/threonine-protein kinase) (FIST) (Androgen receptor-interacting nuclear protein kinase) (ANPK). | |||||
|
HIPK3_MOUSE
|
||||||
| NC score | 0.003646 (rank : 49) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ERH7, O88906, Q9QZR2 | Gene names | Hipk3, Fist3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeodomain-interacting protein kinase 3 (EC 2.7.11.1) (Fas- interacting serine/threonine-protein kinase) (FIST) (Nuclear body- associated kinase 3) (Nbak3) (Androgen receptor-interacting nuclear protein kinase) (ANPK). | |||||
|
HKR3_HUMAN
|
||||||
| NC score | 0.000267 (rank : 50) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10074 | Gene names | HKR3 | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-related zinc finger protein 3 (Protein HKR3). | |||||