Please be patient as the page loads
|
FRAT1_HUMAN
|
||||||
| SwissProt Accessions | Q92837, Q8NE74, Q8TDW9 | Gene names | FRAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FRAT1_HUMAN
|
||||||
| θ value | 4.1775e-114 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q92837, Q8NE74, Q8TDW9 | Gene names | FRAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
FRAT1_MOUSE
|
||||||
| θ value | 1.54831e-84 (rank : 2) | NC score | 0.973794 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70339 | Gene names | Frat1 | |||
|
Domain Architecture |
|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
FRAT2_HUMAN
|
||||||
| θ value | 2.31901e-72 (rank : 3) | NC score | 0.968568 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75474, Q5JTI0, Q8WUL9, Q9BYG2 | Gene names | FRAT2 | |||
|
Domain Architecture |
|
|||||
| Description | GSK-3-binding protein FRAT2 (Frat-2). | |||||
|
FRAT2_MOUSE
|
||||||
| θ value | 1.46607e-42 (rank : 4) | NC score | 0.959844 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K025 | Gene names | Frat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GSK-3-binding protein FRAT2 (Frat-2). | |||||
|
PAWR_HUMAN
|
||||||
| θ value | 0.125558 (rank : 5) | NC score | 0.075258 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 6) | NC score | 0.040349 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
PTN21_MOUSE
|
||||||
| θ value | 0.21417 (rank : 7) | NC score | 0.027989 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
WDR33_HUMAN
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.028772 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.47712 (rank : 9) | NC score | 0.036531 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
PTN21_HUMAN
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.026932 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
STAT2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.025970 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
PAF49_MOUSE
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.076739 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q76KJ5, Q8K0Y9 | Gene names | Cd3eap, Ase1, Paf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein). | |||||
|
COBA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.031459 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
LBX1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.020605 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52954 | Gene names | LBX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor LBX1. | |||||
|
SYNP2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.043529 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
MK15_HUMAN
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.005093 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
COBA2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.036383 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
IG2AS_HUMAN
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.077143 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6U949, Q7Z712, Q9P2W0 | Gene names | IGF2AS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative insulin-like growth factor 2 antisense gene protein (IGF2-AS) (PEG8/IGF2AS protein). | |||||
|
CENPA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.026834 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49450, Q53T74, Q9BVW2 | Gene names | CENPA | |||
|
Domain Architecture |
|
|||||
| Description | Histone H3-like centromeric protein A (Centromere protein A) (CENP-A) (Centromere autoantigen A). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.008485 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
MINK1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.004156 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.019361 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
ZN500_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.002002 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60304, Q96CQ8, Q9BTG0 | Gene names | ZNF500, KIAA0557 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 500. | |||||
|
ZN692_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.006223 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BU19, Q5SRA5, Q5SRA6, Q9HBC9, Q9NW93, Q9NWY6, Q9UF97 | Gene names | ZNF692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.024766 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
SYNP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.039618 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
COGA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.027536 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
MELPH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.020951 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
TM108_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.033172 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6UXF1, Q9BQH1, Q9BW81, Q9C0H3 | Gene names | TMEM108, KIAA1690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.011317 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
VAX1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.008739 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q2NKI2, O88880 | Gene names | Vax1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ventral anterior homeobox 1. | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.027176 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
EMIL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.014768 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NT22, Q76KT4 | Gene names | EMILIN3, C20orf130, EMILIN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMILIN-3 precursor (EMILIN-5) (Elastin microfibril interface-located protein 5) (Elastin microfibril interfacer 5). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.011947 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.019997 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.020808 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.021458 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CEL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.015115 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.030622 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.030752 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
IL4RA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.023823 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |
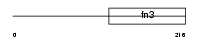
|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
PGES2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.019670 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWM0, Q99J30 | Gene names | Ptges2, Gbf1, Pges2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostaglandin E synthase 2 (EC 5.3.99.3) (Microsomal prostaglandin E synthase 2) (mPGES-2) (GATE-binding factor 1) (GBF-1) [Contains: Prostaglandin E synthase 2 truncated form]. | |||||
|
CO1A2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.022291 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08123, P02464, Q9UEB6, Q9UPH0 | Gene names | COL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
CO8A2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.019244 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P25318, Q68ED0, Q6KAQ4, Q6P1C4 | Gene names | Col8a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(VIII) chain precursor (Endothelial collagen). | |||||
|
DHH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.009734 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61488 | Gene names | Dhh | |||
|
Domain Architecture |
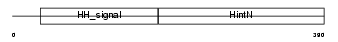
|
|||||
| Description | Desert hedgehog protein precursor (DHH) (HHG-3) [Contains: Desert hedgehog protein N-product; Desert hedgehog protein C-product]. | |||||
|
FZD5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.004674 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13467, Q53R22 | Gene names | FZD5, HFZ5 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-5 precursor (Fz-5) (hFz5) (FzE5). | |||||
|
OSBP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.006980 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.019641 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
FRAT1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.1775e-114 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q92837, Q8NE74, Q8TDW9 | Gene names | FRAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
FRAT1_MOUSE
|
||||||
| NC score | 0.973794 (rank : 2) | θ value | 1.54831e-84 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70339 | Gene names | Frat1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
FRAT2_HUMAN
|
||||||
| NC score | 0.968568 (rank : 3) | θ value | 2.31901e-72 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75474, Q5JTI0, Q8WUL9, Q9BYG2 | Gene names | FRAT2 | |||
|
Domain Architecture |

|
|||||
| Description | GSK-3-binding protein FRAT2 (Frat-2). | |||||
|
FRAT2_MOUSE
|
||||||
| NC score | 0.959844 (rank : 4) | θ value | 1.46607e-42 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K025 | Gene names | Frat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GSK-3-binding protein FRAT2 (Frat-2). | |||||
|
IG2AS_HUMAN
|
||||||
| NC score | 0.077143 (rank : 5) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6U949, Q7Z712, Q9P2W0 | Gene names | IGF2AS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative insulin-like growth factor 2 antisense gene protein (IGF2-AS) (PEG8/IGF2AS protein). | |||||
|
PAF49_MOUSE
|
||||||
| NC score | 0.076739 (rank : 6) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q76KJ5, Q8K0Y9 | Gene names | Cd3eap, Ase1, Paf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein). | |||||
|
PAWR_HUMAN
|
||||||
| NC score | 0.075258 (rank : 7) | θ value | 0.125558 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
SYNP2_HUMAN
|
||||||
| NC score | 0.043529 (rank : 8) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.040349 (rank : 9) | θ value | 0.163984 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
SYNP2_MOUSE
|
||||||
| NC score | 0.039618 (rank : 10) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.036531 (rank : 11) | θ value | 0.47712 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
COBA2_MOUSE
|
||||||
| NC score | 0.036383 (rank : 12) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
TM108_HUMAN
|
||||||
| NC score | 0.033172 (rank : 13) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6UXF1, Q9BQH1, Q9BW81, Q9C0H3 | Gene names | TMEM108, KIAA1690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
COBA1_HUMAN
|
||||||
| NC score | 0.031459 (rank : 14) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.030752 (rank : 15) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.030622 (rank : 16) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
WDR33_HUMAN
|
||||||
| NC score | 0.028772 (rank : 17) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
PTN21_MOUSE
|
||||||
| NC score | 0.027989 (rank : 18) | θ value | 0.21417 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
COGA1_HUMAN
|
||||||
| NC score | 0.027536 (rank : 19) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.027176 (rank : 20) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
PTN21_HUMAN
|
||||||
| NC score | 0.026932 (rank : 21) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
CENPA_HUMAN
|
||||||
| NC score | 0.026834 (rank : 22) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49450, Q53T74, Q9BVW2 | Gene names | CENPA | |||
|
Domain Architecture |

|
|||||
| Description | Histone H3-like centromeric protein A (Centromere protein A) (CENP-A) (Centromere autoantigen A). | |||||
|
STAT2_MOUSE
|
||||||
| NC score | 0.025970 (rank : 23) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.024766 (rank : 24) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
IL4RA_HUMAN
|
||||||
| NC score | 0.023823 (rank : 25) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
CO1A2_HUMAN
|
||||||
| NC score | 0.022291 (rank : 26) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08123, P02464, Q9UEB6, Q9UPH0 | Gene names | COL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.021458 (rank : 27) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
MELPH_HUMAN
|
||||||
| NC score | 0.020951 (rank : 28) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.020808 (rank : 29) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
LBX1_HUMAN
|
||||||
| NC score | 0.020605 (rank : 30) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52954 | Gene names | LBX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor LBX1. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.019997 (rank : 31) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PGES2_MOUSE
|
||||||
| NC score | 0.019670 (rank : 32) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWM0, Q99J30 | Gene names | Ptges2, Gbf1, Pges2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostaglandin E synthase 2 (EC 5.3.99.3) (Microsomal prostaglandin E synthase 2) (mPGES-2) (GATE-binding factor 1) (GBF-1) [Contains: Prostaglandin E synthase 2 truncated form]. | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.019641 (rank : 33) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.019361 (rank : 34) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
CO8A2_MOUSE
|
||||||
| NC score | 0.019244 (rank : 35) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P25318, Q68ED0, Q6KAQ4, Q6P1C4 | Gene names | Col8a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(VIII) chain precursor (Endothelial collagen). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.015115 (rank : 36) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
EMIL3_HUMAN
|
||||||
| NC score | 0.014768 (rank : 37) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NT22, Q76KT4 | Gene names | EMILIN3, C20orf130, EMILIN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMILIN-3 precursor (EMILIN-5) (Elastin microfibril interface-located protein 5) (Elastin microfibril interfacer 5). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.011947 (rank : 38) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.011317 (rank : 39) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
DHH_MOUSE
|
||||||
| NC score | 0.009734 (rank : 40) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61488 | Gene names | Dhh | |||
|
Domain Architecture |

|
|||||
| Description | Desert hedgehog protein precursor (DHH) (HHG-3) [Contains: Desert hedgehog protein N-product; Desert hedgehog protein C-product]. | |||||
|
VAX1_MOUSE
|
||||||
| NC score | 0.008739 (rank : 41) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q2NKI2, O88880 | Gene names | Vax1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ventral anterior homeobox 1. | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.008485 (rank : 42) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
OSBP1_HUMAN
|
||||||
| NC score | 0.006980 (rank : 43) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
|
ZN692_HUMAN
|
||||||
| NC score | 0.006223 (rank : 44) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BU19, Q5SRA5, Q5SRA6, Q9HBC9, Q9NW93, Q9NWY6, Q9UF97 | Gene names | ZNF692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
MK15_HUMAN
|
||||||
| NC score | 0.005093 (rank : 45) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
FZD5_HUMAN
|
||||||
| NC score | 0.004674 (rank : 46) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13467, Q53R22 | Gene names | FZD5, HFZ5 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-5 precursor (Fz-5) (hFz5) (FzE5). | |||||
|
MINK1_MOUSE
|
||||||
| NC score | 0.004156 (rank : 47) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
ZN500_HUMAN
|
||||||
| NC score | 0.002002 (rank : 48) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60304, Q96CQ8, Q9BTG0 | Gene names | ZNF500, KIAA0557 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 500. | |||||