Please be patient as the page loads
|
MELPH_HUMAN
|
||||||
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MELPH_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
MELPH_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.939626 (rank : 2) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
MYRIP_MOUSE
|
||||||
| θ value | 3.37985e-39 (rank : 3) | NC score | 0.740835 (rank : 3) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 6.59618e-35 (rank : 4) | NC score | 0.720170 (rank : 4) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
SYTL4_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 5) | NC score | 0.349531 (rank : 8) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R0Q1, Q8R321, Q9R0Q0 | Gene names | Sytl4, Slp4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYTL5_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 6) | NC score | 0.354411 (rank : 6) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
SYTL3_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 7) | NC score | 0.390502 (rank : 5) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
SYTL4_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 8) | NC score | 0.329866 (rank : 9) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYTL5_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 9) | NC score | 0.352325 (rank : 7) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
SYTL2_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 10) | NC score | 0.263873 (rank : 10) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 11) | NC score | 0.035302 (rank : 116) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 12) | NC score | 0.079616 (rank : 57) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 13) | NC score | 0.178130 (rank : 14) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
LPIN1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 14) | NC score | 0.056689 (rank : 87) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZP3, Q9CQI2, Q9JLG6 | Gene names | Lpin1, Fld | |||
|
Domain Architecture |
|
|||||
| Description | Lipin-1 (Fatty liver dystrophy protein). | |||||
|
RP3A_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 15) | NC score | 0.174438 (rank : 15) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 16) | NC score | 0.118844 (rank : 17) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF10_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 17) | NC score | 0.117029 (rank : 18) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 18) | NC score | 0.034391 (rank : 119) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 19) | NC score | 0.062928 (rank : 74) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TPR_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 20) | NC score | 0.035094 (rank : 117) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
SYTL1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.243296 (rank : 12) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
SLC2B_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 22) | NC score | 0.143880 (rank : 16) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NEV8, Q9Y4D6 | Gene names | EXPH5, KIAA0624, SLAC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Slp homolog lacking C2 domains b (Exophilin-5). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.056152 (rank : 89) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 24) | NC score | 0.249928 (rank : 11) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.080810 (rank : 55) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 26) | NC score | 0.068332 (rank : 67) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
HDAC5_MOUSE
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.033329 (rank : 120) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.078785 (rank : 58) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
TBCD2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.044279 (rank : 104) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
CMGA_MOUSE
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.061992 (rank : 75) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
K1H2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.026470 (rank : 136) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62168 | Gene names | Krtha2, Hka2, Krt1-2 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.067378 (rank : 69) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.076431 (rank : 61) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.061844 (rank : 76) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.035675 (rank : 114) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
FGD2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.083700 (rank : 52) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BY35, O88841, Q7TSE3, Q8VDH4 | Gene names | Fgd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2. | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.035608 (rank : 115) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.076302 (rank : 62) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FGD2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.083144 (rank : 53) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z6J4, Q5T8I1, Q6P6A8, Q6ZNL5, Q8IZ32, Q8N868, Q9H7M2 | Gene names | FGD2, ZFYVE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2 (Zinc finger FYVE domain-containing protein 4). | |||||
|
FGD4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.072728 (rank : 65) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
ZN211_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.000151 (rank : 195) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13398 | Gene names | ZNF211 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 211 (Zinc finger protein C2H2-25). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.012925 (rank : 176) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
ABCF2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.020574 (rank : 160) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LE6, Q3UA24, Q8C1S5, Q9JL48 | Gene names | Abcf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 2. | |||||
|
ATF6B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.047710 (rank : 96) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99941, Q13269, Q14343, Q14345, Q99635, Q99637, Q9H3V9, Q9H3W1, Q9NPL0 | Gene names | CREBL1, G13 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp) (Protein G13). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.049036 (rank : 95) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.034906 (rank : 118) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
HORN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.047481 (rank : 97) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.060438 (rank : 80) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.061003 (rank : 79) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.027370 (rank : 133) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
STX18_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.038909 (rank : 112) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P2W9, Q596L3, Q5TZP5 | Gene names | STX18, GIG9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-18 (Growth-inhibiting gene 9 protein). | |||||
|
ATF6B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.046591 (rank : 99) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
CDKL5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.004433 (rank : 192) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.027694 (rank : 131) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.042181 (rank : 107) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
PKHF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.063784 (rank : 72) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TB82, Q99M16 | Gene names | Plekhf1, Lapf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.008648 (rank : 184) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.060128 (rank : 81) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.011600 (rank : 179) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
SPY2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.039959 (rank : 110) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43597, Q5T6Z7 | Gene names | SPRY2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPY2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.040315 (rank : 109) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXV8, Q9WUQ9 | Gene names | Spry2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
B3A2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.012385 (rank : 177) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04920, Q45EY5, Q969L3 | Gene names | SLC4A2, AE2, EPB3L1, HKB3, MPB3L | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 2 (Non-erythroid band 3-like protein) (AE2 anion exchanger) (Solute carrier family 4 member 2) (BND3L). | |||||
|
KRHB3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.022215 (rank : 149) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P78385, Q9NSB3 | Gene names | KRTHB3 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb3 (Hair keratin, type II Hb3). | |||||
|
LMNB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.029091 (rank : 128) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P14733, Q61791 | Gene names | Lmnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
MYCL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.022230 (rank : 148) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.038921 (rank : 111) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
ZFYV1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.046475 (rank : 100) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HBF4, Q8WYX7, Q96K57, Q9BXP9, Q9HCI3 | Gene names | ZFYVE1, DFCP1, KIAA1589, TAFF1, ZNFN2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1 (Double FYVE-containing protein 1) (Tandem FYVE fingers-1) (SR3). | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.029124 (rank : 127) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
CENPB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.033158 (rank : 121) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
EVX1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.006184 (rank : 186) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23683 | Gene names | Evx1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox even-skipped homolog protein 1 (EVX-1). | |||||
|
FBLI1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.009910 (rank : 183) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q71FD7, Q99J35 | Gene names | Fblim1, Cal | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-binding LIM protein 1 (CSX-associated LIM). | |||||
|
FGD4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.063893 (rank : 71) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
JIP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.027129 (rank : 135) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
K1H2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.023042 (rank : 146) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14532 | Gene names | KRTHA2, HHA2, HKA2 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
LMNB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.030360 (rank : 125) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
MPP10_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.028727 (rank : 130) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.025373 (rank : 138) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PKHF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.056001 (rank : 90) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96S99, Q96K11, Q9BUB9 | Gene names | PLEKHF1, APPD, LAPF, ZFYVE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (PH and FYVE domain-containing protein 1) (Phafin-1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains) (Apoptosis-inducing protein) (Zinc finger FYVE domain-containing protein 15). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.016648 (rank : 171) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.049691 (rank : 94) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ZFYV1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.045514 (rank : 101) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q810J8 | Gene names | Zfyve1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1. | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.024186 (rank : 141) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
FRAT1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.020951 (rank : 155) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92837, Q8NE74, Q8TDW9 | Gene names | FRAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.031234 (rank : 123) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.028907 (rank : 129) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
PCDA5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.004570 (rank : 191) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5H7, O75284, Q8N4R3 | Gene names | PCDHA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 5 precursor (PCDH-alpha5). | |||||
|
PCDA7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.004634 (rank : 190) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UN72, O75282 | Gene names | PCDHA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 7 precursor (PCDH-alpha7). | |||||
|
SNPC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.027223 (rank : 134) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13487, Q13486 | Gene names | SNAPC2, SNAP45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2) (Proximal sequence element-binding transcription factor subunit delta) (PSE- binding factor subunit delta) (PTF subunit delta). | |||||
|
SYNP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.019488 (rank : 163) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.005432 (rank : 188) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
ZN275_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | -0.000370 (rank : 197) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NSD4 | Gene names | ZNF275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 275. | |||||
|
ADA2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | -0.000942 (rank : 198) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08913, Q2XN99, Q86TH8, Q9BZK1 | Gene names | ADRA2A, ADRA2R, ADRAR | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2A adrenergic receptor (Alpha-2A adrenoceptor) (Alpha-2A adrenoreceptor) (Alpha-2AAR) (Alpha-2 adrenergic receptor subtype C10). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.013079 (rank : 175) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ATX3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.021610 (rank : 152) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54252, O15284, O15285, O15286, Q8N189, Q96TC3, Q96TC4, Q9H3N0 | Gene names | ATXN3, ATX3, MJD, MJD1, SCA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-3 (EC 3.4.22.-) (Machado-Joseph disease protein 1) (Spinocerebellar ataxia type 3 protein). | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.023117 (rank : 145) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
CLMN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.011809 (rank : 178) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
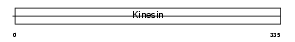
KIF9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.005739 (rank : 187) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HAQ2, Q86Z28, Q9H8A4 | Gene names | KIF9 | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF9. | |||||
|
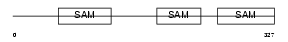
LIPA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.018696 (rank : 167) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75335, O94971 | Gene names | PPFIA4, KIAA0897 | |||
|
Domain Architecture |
|
|||||
| Description | Liprin-alpha-4 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-4) (PTPRF-interacting protein alpha-4). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.026192 (rank : 137) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
NEK3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.000881 (rank : 194) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R0A5, Q9Z0X9 | Gene names | Nek3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek3 (EC 2.7.11.1) (NimA-related protein kinase 3). | |||||
|
REQU_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.044739 (rank : 103) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.045087 (rank : 102) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
SCG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.032757 (rank : 122) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P16014 | Gene names | Chgb, Scg-1, Scg1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB). | |||||
|
SGOL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.018820 (rank : 166) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q562F6, Q53RR9, Q53T20, Q86XY4, Q8IWK2, Q8IZK1, Q8N1Q5, Q96LQ3 | Gene names | SGOL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2 (Tripin). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.008348 (rank : 185) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.038659 (rank : 113) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.209376 (rank : 13) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
TLN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.015990 (rank : 173) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.016114 (rank : 172) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TPM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.020670 (rank : 158) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
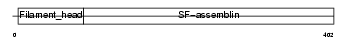
AINX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.020581 (rank : 159) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q16352, Q9BRC5 | Gene names | INA | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-internexin (Alpha-Inx) (66 kDa neurofilament protein) (Neurofilament-66) (NF-66). | |||||
|
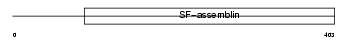
AINX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.020899 (rank : 156) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46660, Q61958, Q8VCW5 | Gene names | Ina | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-internexin (Alpha-Inx) (66 kDa neurofilament protein) (Neurofilament-66) (NF-66). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.025063 (rank : 139) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
ARI5B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.015589 (rank : 174) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.040460 (rank : 108) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.023735 (rank : 143) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.052772 (rank : 91) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.021075 (rank : 154) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GP73_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.030681 (rank : 124) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91XA2 | Gene names | Golph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
KLF10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.000016 (rank : 196) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13118 | Gene names | KLF10, TIEG, TIEG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 10 (Transforming growth factor-beta-inducible early growth response protein 1) (TGFB-inducible early growth response protein 1) (TIEG-1) (EGR-alpha). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.010437 (rank : 182) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.022991 (rank : 147) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYO5B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.023559 (rank : 144) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.043183 (rank : 105) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.021567 (rank : 153) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.042228 (rank : 106) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
ARHG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.020782 (rank : 157) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92888, O00513, Q8N4J4, Q96BF4, Q96F17, Q9BSB1 | Gene names | ARHGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (p115-RhoGEF) (p115RhoGEF) (115 kDa guanine nucleotide exchange factor) (Sub1.5). | |||||
|
CAD15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.003023 (rank : 193) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55291 | Gene names | CDH15, CDH14, CDH3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-15 precursor (Muscle-cadherin) (M-cadherin) (Cadherin-14). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.022198 (rank : 150) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
HDAC5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.024037 (rank : 142) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
K1C19_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.019209 (rank : 164) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19001 | Gene names | Krt19, Krt1-19 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 19 (Cytokeratin-19) (CK-19) (Keratin-19) (K19). | |||||
|
KRHB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.020200 (rank : 162) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14533, Q14846, Q16274, Q8WU52, Q9BR74 | Gene names | KRTHB1, MLN137 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb1 (Hair keratin, type II Hb1) (ghHKb1) (ghHb1) (MLN 137). | |||||
|
KRHB6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.020245 (rank : 161) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43790, P78387 | Gene names | KRTHB6 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6) (ghHb6). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.046870 (rank : 98) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.029602 (rank : 126) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.016982 (rank : 168) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.004802 (rank : 189) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PK3C3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.010686 (rank : 181) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PK3C3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.010690 (rank : 180) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
SACS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.016736 (rank : 169) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.016734 (rank : 170) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.024548 (rank : 140) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.027609 (rank : 132) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.019204 (rank : 165) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
ZMY15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.021784 (rank : 151) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H091 | Gene names | ZMYND15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 15. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.050279 (rank : 93) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.058011 (rank : 85) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DOC2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.113938 (rank : 21) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
|
DOC2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.114361 (rank : 20) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TNF0 | Gene names | Doc2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha). | |||||
|
DOC2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.113805 (rank : 22) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14184 | Gene names | DOC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
DOC2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.114900 (rank : 19) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70169, Q6NXK3 | Gene names | Doc2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
DOC2G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.111640 (rank : 23) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ESN1, Q6P8R8 | Gene names | Doc2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein gamma (Doc2-gamma). | |||||
|
FA62A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.079754 (rank : 56) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
FA62A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.077631 (rank : 59) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
FGD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.051139 (rank : 92) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
P3C2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.073234 (rank : 64) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P3C2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.075291 (rank : 63) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
P3C2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.069092 (rank : 66) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.092397 (rank : 38) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.087522 (rank : 47) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RASA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.058500 (rank : 83) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.056825 (rank : 86) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.081528 (rank : 54) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
RIMS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.094331 (rank : 31) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
RIMS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.091582 (rank : 39) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
RIMS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.061165 (rank : 77) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJD0, Q92511 | Gene names | RIMS3, KIAA0237 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
RIMS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.063689 (rank : 73) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80U57 | Gene names | Rims3, Kiaa0237 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
RIMS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.066854 (rank : 70) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H426, Q5JWT7 | Gene names | RIMS4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
RIMS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.067911 (rank : 68) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60191 | Gene names | Rims4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
SYT10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.088877 (rank : 40) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6XYQ8 | Gene names | SYT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
SYT10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.086318 (rank : 51) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0N4 | Gene names | Syt10 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
SYT11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.087856 (rank : 46) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BT88, Q14998, Q68CT5, Q8IXU3, Q96SU2 | Gene names | SYT11, KIAA0080 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
SYT11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.087893 (rank : 45) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0N3 | Gene names | Syt11 | |||
|
Domain Architecture |
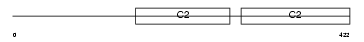
|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
SYT12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.093138 (rank : 35) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IV01 | Gene names | SYT12 | |||
|
Domain Architecture |
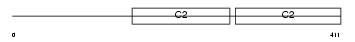
|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYT12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.093534 (rank : 33) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q920N7 | Gene names | Syt12 | |||
|
Domain Architecture |
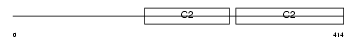
|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYT13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.058235 (rank : 84) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7L8C5, Q9BQS3, Q9H041, Q9P2C0 | Gene names | SYT13, KIAA1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
SYT13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.056180 (rank : 88) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EQT6, Q6ZPR2, Q8BRK6 | Gene names | Syt13, Kiaa1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
SYT14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.061163 (rank : 78) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NB59, Q5THX7, Q707N3, Q707N4, Q707N5, Q707N6, Q707N7 | Gene names | SYT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
SYT14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.058993 (rank : 82) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TN84 | Gene names | Syt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
SYT15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.086716 (rank : 50) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BQS2, Q7Z439, Q7Z440 | Gene names | SYT15 | |||
|
Domain Architecture |
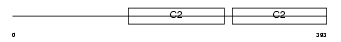
|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV) (Chr10Syt). | |||||
|
SYT15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.076508 (rank : 60) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C6N3, Q7TN81, Q7TN82, Q8C999 | Gene names | Syt15 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV). | |||||
|
SYT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.093127 (rank : 36) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P21579 | Gene names | SYT1, SYT | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.093127 (rank : 37) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46096 | Gene names | Syt1 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.096358 (rank : 28) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N9I0, Q496K5, Q8NBE5 | Gene names | SYT2 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.096534 (rank : 27) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46097, Q8R0E1 | Gene names | Syt2 | |||
|
Domain Architecture |
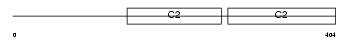
|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.094473 (rank : 30) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.094099 (rank : 32) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.088720 (rank : 42) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H2B2, Q9P2K4 | Gene names | SYT4, KIAA1342 | |||
|
Domain Architecture |
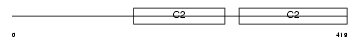
|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
SYT4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.088433 (rank : 44) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P40749, Q3UFC1, Q8BGH3, Q8BRL6 | Gene names | Syt4, Syt3 | |||
|
Domain Architecture |
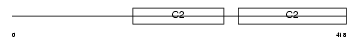
|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
SYT5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.097647 (rank : 25) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00445, Q86X72 | Gene names | SYT5 | |||
|
Domain Architecture |
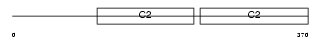
|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV). | |||||
|
SYT5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.099839 (rank : 24) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0N5 | Gene names | Syt5, Syt9 | |||
|
Domain Architecture |
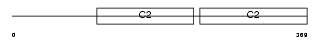
|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV) (Synaptotagmin IX). | |||||
|
SYT6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.088686 (rank : 43) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T7P8 | Gene names | SYT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
SYT6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.088730 (rank : 41) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0N8 | Gene names | Syt6 | |||
|
Domain Architecture |
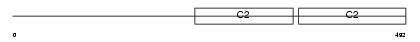
|
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
SYT7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.097013 (rank : 26) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43581 | Gene names | SYT7 | |||
|
Domain Architecture |
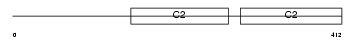
|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
SYT7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.094800 (rank : 29) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0N7 | Gene names | Syt7 | |||
|
Domain Architecture |
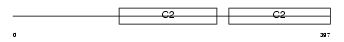
|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
SYT8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.093363 (rank : 34) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0N6, Q64366, Q8C6E7 | Gene names | Syt8 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-8 (Synaptotagmin VIII) (SytVIII). | |||||
|
SYT9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 197) | NC score | 0.086987 (rank : 48) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86SS6 | Gene names | SYT9 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX). | |||||
|
SYT9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 198) | NC score | 0.086855 (rank : 49) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0N9, Q8C280 | Gene names | Syt9, Syt5 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX) (Synaptotagmin V). | |||||
|
MELPH_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
MELPH_MOUSE
|
||||||
| NC score | 0.939626 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
MYRIP_MOUSE
|
||||||
| NC score | 0.740835 (rank : 3) | θ value | 3.37985e-39 (rank : 3) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.720170 (rank : 4) | θ value | 6.59618e-35 (rank : 4) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
SYTL3_MOUSE
|
||||||
| NC score | 0.390502 (rank : 5) | θ value | 4.30538e-10 (rank : 7) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
SYTL5_HUMAN
|
||||||
| NC score | 0.354411 (rank : 6) | θ value | 8.67504e-11 (rank : 6) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
SYTL5_MOUSE
|
||||||
| NC score | 0.352325 (rank : 7) | θ value | 6.21693e-09 (rank : 9) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
SYTL4_MOUSE
|
||||||
| NC score | 0.349531 (rank : 8) | θ value | 8.38298e-14 (rank : 5) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R0Q1, Q8R321, Q9R0Q0 | Gene names | Sytl4, Slp4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYTL4_HUMAN
|
||||||
| NC score | 0.329866 (rank : 9) | θ value | 2.13673e-09 (rank : 8) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYTL2_MOUSE
|
||||||
| NC score | 0.263873 (rank : 10) | θ value | 0.00035302 (rank : 10) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.249928 (rank : 11) | θ value | 0.0736092 (rank : 24) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
SYTL1_HUMAN
|
||||||
| NC score | 0.243296 (rank : 12) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.209376 (rank : 13) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.178130 (rank : 14) | θ value | 0.00228821 (rank : 13) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
RP3A_HUMAN
|
||||||
| NC score | 0.174438 (rank : 15) | θ value | 0.00509761 (rank : 15) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SLC2B_HUMAN
|
||||||
| NC score | 0.143880 (rank : 16) | θ value | 0.0563607 (rank : 22) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NEV8, Q9Y4D6 | Gene names | EXPH5, KIAA0624, SLAC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Slp homolog lacking C2 domains b (Exophilin-5). | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.118844 (rank : 17) | θ value | 0.00665767 (rank : 16) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.117029 (rank : 18) | θ value | 0.00869519 (rank : 17) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
DOC2B_MOUSE
|
||||||
| NC score | 0.114900 (rank : 19) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70169, Q6NXK3 | Gene names | Doc2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
DOC2A_MOUSE
|
||||||
| NC score | 0.114361 (rank : 20) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TNF0 | Gene names | Doc2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha). | |||||
|
DOC2A_HUMAN
|
||||||
| NC score | 0.113938 (rank : 21) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
|
DOC2B_HUMAN
|
||||||
| NC score | 0.113805 (rank : 22) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14184 | Gene names | DOC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
DOC2G_MOUSE
|
||||||
| NC score | 0.111640 (rank : 23) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ESN1, Q6P8R8 | Gene names | Doc2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein gamma (Doc2-gamma). | |||||
|
SYT5_MOUSE
|
||||||
| NC score | 0.099839 (rank : 24) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0N5 | Gene names | Syt5, Syt9 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV) (Synaptotagmin IX). | |||||
|
SYT5_HUMAN
|
||||||
| NC score | 0.097647 (rank : 25) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00445, Q86X72 | Gene names | SYT5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV). | |||||
|
SYT7_HUMAN
|
||||||
| NC score | 0.097013 (rank : 26) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43581 | Gene names | SYT7 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
SYT2_MOUSE
|
||||||
| NC score | 0.096534 (rank : 27) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46097, Q8R0E1 | Gene names | Syt2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT2_HUMAN
|
||||||
| NC score | 0.096358 (rank : 28) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N9I0, Q496K5, Q8NBE5 | Gene names | SYT2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT7_MOUSE
|
||||||
| NC score | 0.094800 (rank : 29) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0N7 | Gene names | Syt7 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
SYT3_HUMAN
|
||||||
| NC score | 0.094473 (rank : 30) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
RIMS2_HUMAN
|
||||||
| NC score | 0.094331 (rank : 31) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
SYT3_MOUSE
|
||||||
| NC score | 0.094099 (rank : 32) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT12_MOUSE
|
||||||
| NC score | 0.093534 (rank : 33) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q920N7 | Gene names | Syt12 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYT8_MOUSE
|
||||||
| NC score | 0.093363 (rank : 34) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0N6, Q64366, Q8C6E7 | Gene names | Syt8 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-8 (Synaptotagmin VIII) (SytVIII). | |||||
|
SYT12_HUMAN
|
||||||
| NC score | 0.093138 (rank : 35) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IV01 | Gene names | SYT12 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYT1_HUMAN
|
||||||
| NC score | 0.093127 (rank : 36) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P21579 | Gene names | SYT1, SYT | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT1_MOUSE
|
||||||
| NC score | 0.093127 (rank : 37) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46096 | Gene names | Syt1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.092397 (rank : 38) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RIMS2_MOUSE
|
||||||
| NC score | 0.091582 (rank : 39) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
SYT10_HUMAN
|
||||||
| NC score | 0.088877 (rank : 40) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6XYQ8 | Gene names | SYT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
SYT6_MOUSE
|
||||||
| NC score | 0.088730 (rank : 41) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0N8 | Gene names | Syt6 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
SYT4_HUMAN
|
||||||
| NC score | 0.088720 (rank : 42) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H2B2, Q9P2K4 | Gene names | SYT4, KIAA1342 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
SYT6_HUMAN
|
||||||
| NC score | 0.088686 (rank : 43) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T7P8 | Gene names | SYT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
SYT4_MOUSE
|
||||||
| NC score | 0.088433 (rank : 44) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P40749, Q3UFC1, Q8BGH3, Q8BRL6 | Gene names | Syt4, Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
SYT11_MOUSE
|
||||||
| NC score | 0.087893 (rank : 45) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0N3 | Gene names | Syt11 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
SYT11_HUMAN
|
||||||
| NC score | 0.087856 (rank : 46) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BT88, Q14998, Q68CT5, Q8IXU3, Q96SU2 | Gene names | SYT11, KIAA0080 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.087522 (rank : 47) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SYT9_HUMAN
|
||||||
| NC score | 0.086987 (rank : 48) | θ value | θ > 10 (rank : 197) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86SS6 | Gene names | SYT9 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX). | |||||
|
SYT9_MOUSE
|
||||||
| NC score | 0.086855 (rank : 49) | θ value | θ > 10 (rank : 198) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0N9, Q8C280 | Gene names | Syt9, Syt5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX) (Synaptotagmin V). | |||||
|
SYT15_HUMAN
|
||||||
| NC score | 0.086716 (rank : 50) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BQS2, Q7Z439, Q7Z440 | Gene names | SYT15 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV) (Chr10Syt). | |||||
|
SYT10_MOUSE
|
||||||
| NC score | 0.086318 (rank : 51) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0N4 | Gene names | Syt10 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
FGD2_MOUSE
|
||||||
| NC score | 0.083700 (rank : 52) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BY35, O88841, Q7TSE3, Q8VDH4 | Gene names | Fgd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2. | |||||
|
FGD2_HUMAN
|
||||||
| NC score | 0.083144 (rank : 53) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z6J4, Q5T8I1, Q6P6A8, Q6ZNL5, Q8IZ32, Q8N868, Q9H7M2 | Gene names | FGD2, ZFYVE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2 (Zinc finger FYVE domain-containing protein 4). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.081528 (rank : 54) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.080810 (rank : 55) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
FA62A_HUMAN
|
||||||
| NC score | 0.079754 (rank : 56) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.079616 (rank : 57) | θ value | 0.00228821 (rank : 12) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.078785 (rank : 58) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
FA62A_MOUSE
|
||||||
| NC score | 0.077631 (rank : 59) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
SYT15_MOUSE
|
||||||
| NC score | 0.076508 (rank : 60) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C6N3, Q7TN81, Q7TN82, Q8C999 | Gene names | Syt15 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV). | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.076431 (rank : 61) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.076302 (rank : 62) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
P3C2A_MOUSE
|
||||||
| NC score | 0.075291 (rank : 63) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
P3C2A_HUMAN
|
||||||
| NC score | 0.073234 (rank : 64) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
FGD4_HUMAN
|
||||||
| NC score | 0.072728 (rank : 65) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
P3C2B_HUMAN
|
||||||
| NC score | 0.069092 (rank : 66) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.068332 (rank : 67) | θ value | 0.0961366 (rank : 26) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
RIMS4_MOUSE
|
||||||
| NC score | 0.067911 (rank : 68) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60191 | Gene names | Rims4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.067378 (rank : 69) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
RIMS4_HUMAN
|
||||||
| NC score | 0.066854 (rank : 70) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H426, Q5JWT7 | Gene names | RIMS4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
FGD4_MOUSE
|
||||||
| NC score | 0.063893 (rank : 71) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
PKHF1_MOUSE
|
||||||
| NC score | 0.063784 (rank : 72) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TB82, Q99M16 | Gene names | Plekhf1, Lapf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains). | |||||
|
RIMS3_MOUSE
|
||||||
| NC score | 0.063689 (rank : 73) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80U57 | Gene names | Rims3, Kiaa0237 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.062928 (rank : 74) | θ value | 0.0193708 (rank : 19) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
CMGA_MOUSE
|
||||||
| NC score | 0.061992 (rank : 75) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.061844 (rank : 76) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
RIMS3_HUMAN
|
||||||
| NC score | 0.061165 (rank : 77) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJD0, Q92511 | Gene names | RIMS3, KIAA0237 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
SYT14_HUMAN
|
||||||
| NC score | 0.061163 (rank : 78) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NB59, Q5THX7, Q707N3, Q707N4, Q707N5, Q707N6, Q707N7 | Gene names | SYT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.061003 (rank : 79) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.060438 (rank : 80) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.060128 (rank : 81) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SYT14_MOUSE
|
||||||
| NC score | 0.058993 (rank : 82) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TN84 | Gene names | Syt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
RASA2_HUMAN
|
||||||
| NC score | 0.058500 (rank : 83) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
SYT13_HUMAN
|
||||||
| NC score | 0.058235 (rank : 84) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7L8C5, Q9BQS3, Q9H041, Q9P2C0 | Gene names | SYT13, KIAA1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.058011 (rank : 85) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
RASA2_MOUSE
|
||||||
| NC score | 0.056825 (rank : 86) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
LPIN1_MOUSE
|
||||||
| NC score | 0.056689 (rank : 87) | θ value | 0.00390308 (rank : 14) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZP3, Q9CQI2, Q9JLG6 | Gene names | Lpin1, Fld | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-1 (Fatty liver dystrophy protein). | |||||
|
SYT13_MOUSE
|
||||||
| NC score | 0.056180 (rank : 88) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EQT6, Q6ZPR2, Q8BRK6 | Gene names | Syt13, Kiaa1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.056152 (rank : 89) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
PKHF1_HUMAN
|
||||||
| NC score | 0.056001 (rank : 90) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96S99, Q96K11, Q9BUB9 | Gene names | PLEKHF1, APPD, LAPF, ZFYVE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (PH and FYVE domain-containing protein 1) (Phafin-1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains) (Apoptosis-inducing protein) (Zinc finger FYVE domain-containing protein 15). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.052772 (rank : 91) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FGD3_HUMAN
|
||||||
| NC score | 0.051139 (rank : 92) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.050279 (rank : 93) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.049691 (rank : 94) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.049036 (rank : 95) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
ATF6B_HUMAN
|
||||||
| NC score | 0.047710 (rank : 96) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99941, Q13269, Q14343, Q14345, Q99635, Q99637, Q9H3V9, Q9H3W1, Q9NPL0 | Gene names | CREBL1, G13 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp) (Protein G13). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.047481 (rank : 97) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.046870 (rank : 98) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
ATF6B_MOUSE
|
||||||
| NC score | 0.046591 (rank : 99) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
ZFYV1_HUMAN
|
||||||
| NC score | 0.046475 (rank : 100) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HBF4, Q8WYX7, Q96K57, Q9BXP9, Q9HCI3 | Gene names | ZFYVE1, DFCP1, KIAA1589, TAFF1, ZNFN2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1 (Double FYVE-containing protein 1) (Tandem FYVE fingers-1) (SR3). | |||||
|
ZFYV1_MOUSE
|
||||||
| NC score | 0.045514 (rank : 101) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q810J8 | Gene names | Zfyve1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1. | |||||
|
REQU_MOUSE
|
||||||
| NC score | 0.045087 (rank : 102) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_HUMAN
|
||||||
| NC score | 0.044739 (rank : 103) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
TBCD2_HUMAN
|
||||||
| NC score | 0.044279 (rank : 104) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.043183 (rank : 105) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.042228 (rank : 106) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.042181 (rank : 107) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.040460 (rank : 108) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
SPY2_MOUSE
|
||||||
| NC score | 0.040315 (rank : 109) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXV8, Q9WUQ9 | Gene names | Spry2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPY2_HUMAN
|
||||||
| NC score | 0.039959 (rank : 110) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43597, Q5T6Z7 | Gene names | SPRY2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.038921 (rank : 111) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
STX18_HUMAN
|
||||||
| NC score | 0.038909 (rank : 112) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P2W9, Q596L3, Q5TZP5 | Gene names | STX18, GIG9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-18 (Growth-inhibiting gene 9 protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.038659 (rank : 113) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.035675 (rank : 114) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.035608 (rank : 115) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.035302 (rank : 116) | θ value | 0.00175202 (rank : 11) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.035094 (rank : 117) | θ value | 0.0193708 (rank : 20) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.034906 (rank : 118) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.034391 (rank : 119) | θ value | 0.0113563 (rank : 18) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
HDAC5_MOUSE
|
||||||
| NC score | 0.033329 (rank : 120) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
CENPB_HUMAN
|
||||||
| NC score | 0.033158 (rank : 121) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
SCG1_MOUSE
|
||||||
| NC score | 0.032757 (rank : 122) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P16014 | Gene names | Chgb, Scg-1, Scg1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.031234 (rank : 123) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
GP73_MOUSE
|
||||||
| NC score | 0.030681 (rank : 124) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91XA2 | Gene names | Golph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
LMNB1_HUMAN
|
||||||
| NC score | 0.030360 (rank : 125) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.029602 (rank : 126) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.029124 (rank : 127) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
LMNB1_MOUSE
|
||||||
| NC score | 0.029091 (rank : 128) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P14733, Q61791 | Gene names | Lmnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.028907 (rank : 129) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
MPP10_MOUSE
|
||||||
| NC score | 0.028727 (rank : 130) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.027694 (rank : 131) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.027609 (rank : 132) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.027370 (rank : 133) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
SNPC2_HUMAN
|
||||||
| NC score | 0.027223 (rank : 134) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13487, Q13486 | Gene names | SNAPC2, SNAP45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2) (Proximal sequence element-binding transcription factor subunit delta) (PSE- binding factor subunit delta) (PTF subunit delta). | |||||
|
JIP2_HUMAN
|
||||||
| NC score | 0.027129 (rank : 135) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
K1H2_MOUSE
|
||||||
| NC score | 0.026470 (rank : 136) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62168 | Gene names | Krtha2, Hka2, Krt1-2 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.026192 (rank : 137) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.025373 (rank : 138) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.025063 (rank : 139) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.024548 (rank : 140) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.024186 (rank : 141) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
HDAC5_HUMAN
|
||||||
| NC score | 0.024037 (rank : 142) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.023735 (rank : 143) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
MYO5B_HUMAN
|
||||||
| NC score | 0.023559 (rank : 144) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.023117 (rank : 145) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
K1H2_HUMAN
|
||||||
| NC score | 0.023042 (rank : 146) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14532 | Gene names | KRTHA2, HHA2, HKA2 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.022991 (rank : 147) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYCL2_HUMAN
|
||||||
| NC score | 0.022230 (rank : 148) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
KRHB3_HUMAN
|
||||||
| NC score | 0.022215 (rank : 149) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P78385, Q9NSB3 | Gene names | KRTHB3 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb3 (Hair keratin, type II Hb3). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.022198 (rank : 150) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
ZMY15_HUMAN
|
||||||
| NC score | 0.021784 (rank : 151) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H091 | Gene names | ZMYND15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 15. | |||||
|
ATX3_HUMAN
|
||||||
| NC score | 0.021610 (rank : 152) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54252, O15284, O15285, O15286, Q8N189, Q96TC3, Q96TC4, Q9H3N0 | Gene names | ATXN3, ATX3, MJD, MJD1, SCA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-3 (EC 3.4.22.-) (Machado-Joseph disease protein 1) (Spinocerebellar ataxia type 3 protein). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.021567 (rank : 153) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.021075 (rank : 154) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
FRAT1_HUMAN
|
||||||
| NC score | 0.020951 (rank : 155) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92837, Q8NE74, Q8TDW9 | Gene names | FRAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
AINX_MOUSE
|
||||||
| NC score | 0.020899 (rank : 156) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46660, Q61958, Q8VCW5 | Gene names | Ina | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-internexin (Alpha-Inx) (66 kDa neurofilament protein) (Neurofilament-66) (NF-66). | |||||
|
ARHG1_HUMAN
|
||||||
| NC score | 0.020782 (rank : 157) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92888, O00513, Q8N4J4, Q96BF4, Q96F17, Q9BSB1 | Gene names | ARHGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (p115-RhoGEF) (p115RhoGEF) (115 kDa guanine nucleotide exchange factor) (Sub1.5). | |||||
|
TPM1_MOUSE
|
||||||
| NC score | 0.020670 (rank : 158) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
AINX_HUMAN
|
||||||
| NC score | 0.020581 (rank : 159) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q16352, Q9BRC5 | Gene names | INA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-internexin (Alpha-Inx) (66 kDa neurofilament protein) (Neurofilament-66) (NF-66). | |||||
|
ABCF2_MOUSE
|
||||||
| NC score | 0.020574 (rank : 160) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LE6, Q3UA24, Q8C1S5, Q9JL48 | Gene names | Abcf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 2. | |||||
|
KRHB6_HUMAN
|
||||||
| NC score | 0.020245 (rank : 161) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43790, P78387 | Gene names | KRTHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6) (ghHb6). | |||||
|
KRHB1_HUMAN
|
||||||
| NC score | 0.020200 (rank : 162) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14533, Q14846, Q16274, Q8WU52, Q9BR74 | Gene names | KRTHB1, MLN137 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb1 (Hair keratin, type II Hb1) (ghHKb1) (ghHb1) (MLN 137). | |||||
|
SYNP2_MOUSE
|
||||||
| NC score | 0.019488 (rank : 163) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
K1C19_MOUSE
|
||||||
| NC score | 0.019209 (rank : 164) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19001 | Gene names | Krt19, Krt1-19 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 19 (Cytokeratin-19) (CK-19) (Keratin-19) (K19). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.019204 (rank : 165) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
SGOL2_HUMAN
|
||||||
| NC score | 0.018820 (rank : 166) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q562F6, Q53RR9, Q53T20, Q86XY4, Q8IWK2, Q8IZK1, Q8N1Q5, Q96LQ3 | Gene names | SGOL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2 (Tripin). | |||||
|
LIPA4_HUMAN
|
||||||
| NC score | 0.018696 (rank : 167) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75335, O94971 | Gene names | PPFIA4, KIAA0897 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-4 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-4) (PTPRF-interacting protein alpha-4). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.016982 (rank : 168) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
SACS_HUMAN
|
||||||
| NC score | 0.016736 (rank : 169) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_MOUSE
|
||||||
| NC score | 0.016734 (rank : 170) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.016648 (rank : 171) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
TLN1_MOUSE
|
||||||
| NC score | 0.016114 (rank : 172) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_HUMAN
|
||||||
| NC score | 0.015990 (rank : 173) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
ARI5B_MOUSE
|
||||||
| NC score | 0.015589 (rank : 174) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.013079 (rank : 175) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.012925 (rank : 176) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
B3A2_HUMAN
|
||||||
| NC score | 0.012385 (rank : 177) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04920, Q45EY5, Q969L3 | Gene names | SLC4A2, AE2, EPB3L1, HKB3, MPB3L | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 2 (Non-erythroid band 3-like protein) (AE2 anion exchanger) (Solute carrier family 4 member 2) (BND3L). | |||||
|
CLMN_HUMAN
|
||||||
| NC score | 0.011809 (rank : 178) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.011600 (rank : 179) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
PK3C3_MOUSE
|
||||||
| NC score | 0.010690 (rank : 180) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
PK3C3_HUMAN
|
||||||
| NC score | 0.010686 (rank : 181) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.010437 (rank : 182) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
FBLI1_MOUSE
|
||||||
| NC score | 0.009910 (rank : 183) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q71FD7, Q99J35 | Gene names | Fblim1, Cal | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-binding LIM protein 1 (CSX-associated LIM). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.008648 (rank : 184) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.008348 (rank : 185) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
EVX1_MOUSE
|
||||||
| NC score | 0.006184 (rank : 186) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23683 | Gene names | Evx1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox even-skipped homolog protein 1 (EVX-1). | |||||
|
KIF9_HUMAN
|
||||||
| NC score | 0.005739 (rank : 187) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HAQ2, Q86Z28, Q9H8A4 | Gene names | KIF9 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF9. | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.005432 (rank : 188) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.004802 (rank : 189) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PCDA7_HUMAN
|
||||||
| NC score | 0.004634 (rank : 190) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UN72, O75282 | Gene names | PCDHA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 7 precursor (PCDH-alpha7). | |||||
|
PCDA5_HUMAN
|
||||||
| NC score | 0.004570 (rank : 191) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5H7, O75284, Q8N4R3 | Gene names | PCDHA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 5 precursor (PCDH-alpha5). | |||||
|
CDKL5_HUMAN
|
||||||
| NC score | 0.004433 (rank : 192) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
CAD15_HUMAN
|
||||||
| NC score | 0.003023 (rank : 193) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55291 | Gene names | CDH15, CDH14, CDH3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-15 precursor (Muscle-cadherin) (M-cadherin) (Cadherin-14). | |||||
|
NEK3_MOUSE
|
||||||
| NC score | 0.000881 (rank : 194) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R0A5, Q9Z0X9 | Gene names | Nek3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek3 (EC 2.7.11.1) (NimA-related protein kinase 3). | |||||
|
ZN211_HUMAN
|
||||||
| NC score | 0.000151 (rank : 195) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13398 | Gene names | ZNF211 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 211 (Zinc finger protein C2H2-25). | |||||
|
KLF10_HUMAN
|
||||||
| NC score | 0.000016 (rank : 196) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13118 | Gene names | KLF10, TIEG, TIEG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 10 (Transforming growth factor-beta-inducible early growth response protein 1) (TGFB-inducible early growth response protein 1) (TIEG-1) (EGR-alpha). | |||||
|
ZN275_HUMAN
|
||||||
| NC score | -0.000370 (rank : 197) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NSD4 | Gene names | ZNF275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 275. | |||||
|
ADA2A_HUMAN
|
||||||
| NC score | -0.000942 (rank : 198) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08913, Q2XN99, Q86TH8, Q9BZK1 | Gene names | ADRA2A, ADRA2R, ADRAR | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2A adrenergic receptor (Alpha-2A adrenoceptor) (Alpha-2A adrenoreceptor) (Alpha-2AAR) (Alpha-2 adrenergic receptor subtype C10). | |||||