Please be patient as the page loads
|
DOC2A_HUMAN
|
||||||
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DOC2A_HUMAN
|
||||||
| θ value | 3.97187e-173 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
|
DOC2A_MOUSE
|
||||||
| θ value | 1.15563e-172 (rank : 2) | NC score | 0.998683 (rank : 2) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q7TNF0 | Gene names | Doc2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha). | |||||
|
DOC2B_HUMAN
|
||||||
| θ value | 1.48236e-119 (rank : 3) | NC score | 0.989682 (rank : 3) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q14184 | Gene names | DOC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
DOC2B_MOUSE
|
||||||
| θ value | 2.36663e-117 (rank : 4) | NC score | 0.988803 (rank : 4) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | P70169, Q6NXK3 | Gene names | Doc2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
RP3A_HUMAN
|
||||||
| θ value | 5.28454e-109 (rank : 5) | NC score | 0.966335 (rank : 6) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 1.75921e-104 (rank : 6) | NC score | 0.955150 (rank : 7) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
DOC2G_MOUSE
|
||||||
| θ value | 9.69798e-87 (rank : 7) | NC score | 0.974548 (rank : 5) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9ESN1, Q6P8R8 | Gene names | Doc2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein gamma (Doc2-gamma). | |||||
|
SYT4_HUMAN
|
||||||
| θ value | 4.568e-36 (rank : 8) | NC score | 0.911245 (rank : 18) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9H2B2, Q9P2K4 | Gene names | SYT4, KIAA1342 | |||
|
Domain Architecture |
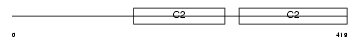
|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
SYT2_MOUSE
|
||||||
| θ value | 1.01765e-35 (rank : 9) | NC score | 0.918232 (rank : 12) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | P46097, Q8R0E1 | Gene names | Syt2 | |||
|
Domain Architecture |
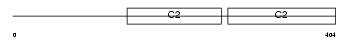
|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT2_HUMAN
|
||||||
| θ value | 1.73584e-35 (rank : 10) | NC score | 0.918155 (rank : 13) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | Q8N9I0, Q496K5, Q8NBE5 | Gene names | SYT2 | |||
|
Domain Architecture |
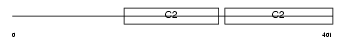
|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT7_MOUSE
|
||||||
| θ value | 5.0505e-35 (rank : 11) | NC score | 0.928090 (rank : 8) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q9R0N7 | Gene names | Syt7 | |||
|
Domain Architecture |
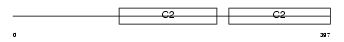
|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
SYT1_HUMAN
|
||||||
| θ value | 8.61488e-35 (rank : 12) | NC score | 0.918293 (rank : 10) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | P21579 | Gene names | SYT1, SYT | |||
|
Domain Architecture |
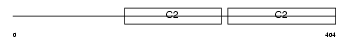
|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT1_MOUSE
|
||||||
| θ value | 8.61488e-35 (rank : 13) | NC score | 0.918271 (rank : 11) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | P46096 | Gene names | Syt1 | |||
|
Domain Architecture |
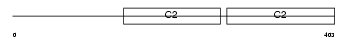
|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT3_HUMAN
|
||||||
| θ value | 8.61488e-35 (rank : 14) | NC score | 0.912066 (rank : 17) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT4_MOUSE
|
||||||
| θ value | 2.50655e-34 (rank : 15) | NC score | 0.907769 (rank : 20) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | P40749, Q3UFC1, Q8BGH3, Q8BRL6 | Gene names | Syt4, Syt3 | |||
|
Domain Architecture |
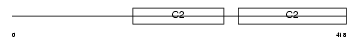
|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
SYT6_MOUSE
|
||||||
| θ value | 2.50655e-34 (rank : 16) | NC score | 0.908291 (rank : 19) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | Q9R0N8 | Gene names | Syt6 | |||
|
Domain Architecture |
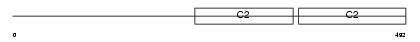
|
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
SYT3_MOUSE
|
||||||
| θ value | 7.29293e-34 (rank : 17) | NC score | 0.912204 (rank : 16) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT6_HUMAN
|
||||||
| θ value | 8.06329e-33 (rank : 18) | NC score | 0.906286 (rank : 21) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | Q5T7P8 | Gene names | SYT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
SYT11_HUMAN
|
||||||
| θ value | 3.06405e-32 (rank : 19) | NC score | 0.900937 (rank : 25) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9BT88, Q14998, Q68CT5, Q8IXU3, Q96SU2 | Gene names | SYT11, KIAA0080 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
SYT11_MOUSE
|
||||||
| θ value | 3.06405e-32 (rank : 20) | NC score | 0.901513 (rank : 24) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q9R0N3 | Gene names | Syt11 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
SYT5_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 21) | NC score | 0.913803 (rank : 14) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | O00445, Q86X72 | Gene names | SYT5 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV). | |||||
|
SYT7_HUMAN
|
||||||
| θ value | 2.86786e-30 (rank : 22) | NC score | 0.926111 (rank : 9) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | O43581 | Gene names | SYT7 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
SYT10_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 23) | NC score | 0.904711 (rank : 22) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q6XYQ8 | Gene names | SYT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
SYT10_MOUSE
|
||||||
| θ value | 4.14116e-29 (rank : 24) | NC score | 0.903191 (rank : 23) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q9R0N4 | Gene names | Syt10 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
SYTL4_HUMAN
|
||||||
| θ value | 5.97985e-28 (rank : 25) | NC score | 0.854962 (rank : 29) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYT9_HUMAN
|
||||||
| θ value | 2.96777e-27 (rank : 26) | NC score | 0.895939 (rank : 26) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q86SS6 | Gene names | SYT9 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX). | |||||
|
SYTL4_MOUSE
|
||||||
| θ value | 6.61148e-27 (rank : 27) | NC score | 0.849046 (rank : 32) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9R0Q1, Q8R321, Q9R0Q0 | Gene names | Sytl4, Slp4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYT9_MOUSE
|
||||||
| θ value | 8.63488e-27 (rank : 28) | NC score | 0.895578 (rank : 27) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | Q9R0N9, Q8C280 | Gene names | Syt9, Syt5 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX) (Synaptotagmin V). | |||||
|
SYT5_MOUSE
|
||||||
| θ value | 6.18819e-25 (rank : 29) | NC score | 0.912346 (rank : 15) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | Q9R0N5 | Gene names | Syt5, Syt9 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV) (Synaptotagmin IX). | |||||
|
SYTL5_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 30) | NC score | 0.820143 (rank : 35) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
SYT15_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 31) | NC score | 0.854137 (rank : 30) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9BQS2, Q7Z439, Q7Z440 | Gene names | SYT15 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV) (Chr10Syt). | |||||
|
SYT12_MOUSE
|
||||||
| θ value | 3.51386e-20 (rank : 32) | NC score | 0.855576 (rank : 28) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q920N7 | Gene names | Syt12 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYT12_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 33) | NC score | 0.853177 (rank : 31) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q8IV01 | Gene names | SYT12 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYT15_MOUSE
|
||||||
| θ value | 1.13037e-18 (rank : 34) | NC score | 0.830647 (rank : 34) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8C6N3, Q7TN81, Q7TN82, Q8C999 | Gene names | Syt15 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV). | |||||
|
SYTL1_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 35) | NC score | 0.811403 (rank : 36) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
SYTL5_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 36) | NC score | 0.810765 (rank : 37) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 37) | NC score | 0.546740 (rank : 57) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SYTL3_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 38) | NC score | 0.781794 (rank : 40) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 39) | NC score | 0.796487 (rank : 38) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.16975e-15 (rank : 40) | NC score | 0.516303 (rank : 58) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SYT14_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 41) | NC score | 0.758356 (rank : 42) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7TN84 | Gene names | Syt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
KPCB_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 42) | NC score | 0.227823 (rank : 110) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 127 | |
| SwissProt Accessions | P68404, P04410, P04411 | Gene names | Prkcb1, Pkcb, Prkcb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
SYTL2_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 43) | NC score | 0.791535 (rank : 39) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
KPCB_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 44) | NC score | 0.224895 (rank : 111) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | P05771, O43744, P05127, Q15138, Q93060, Q9UE49, Q9UE50, Q9UEH8, Q9UJ30, Q9UJ33 | Gene names | PRKCB1, PKCB, PRKCB | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
KPCA_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 45) | NC score | 0.228334 (rank : 109) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | P17252, Q15137, Q96RE4 | Gene names | PRKCA, PKCA, PRKACA | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
RASL2_HUMAN
|
||||||
| θ value | 7.58209e-15 (rank : 46) | NC score | 0.672344 (rank : 48) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | O43374, O60286, Q86UW3, Q96QU0 | Gene names | RASA4, CAPRI, GAPL, KIAA0538 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2) (Calcium-promoted Ras inactivator). | |||||
|
KPCA_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 47) | NC score | 0.224336 (rank : 112) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | P20444 | Gene names | Prkca, Pkca | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
SYT14_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 48) | NC score | 0.753649 (rank : 43) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NB59, Q5THX7, Q707N3, Q707N4, Q707N5, Q707N6, Q707N7 | Gene names | SYT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 49) | NC score | 0.777139 (rank : 41) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
KPCG_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 50) | NC score | 0.243081 (rank : 106) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 127 | |
| SwissProt Accessions | P63318, P05697 | Gene names | Prkcg, Pkcc, Pkcg, Prkcc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
P3C2B_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 51) | NC score | 0.499957 (rank : 61) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
SYT8_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 52) | NC score | 0.837063 (rank : 33) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9R0N6, Q64366, Q8C6E7 | Gene names | Syt8 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-8 (Synaptotagmin VIII) (SytVIII). | |||||
|
KPCG_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 53) | NC score | 0.243175 (rank : 105) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1126 | Shared Neighborhood Hits | 127 | |
| SwissProt Accessions | P05129 | Gene names | PRKCG, PKCG | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
RIMS2_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 54) | NC score | 0.597378 (rank : 49) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
RIMS2_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 55) | NC score | 0.590733 (rank : 51) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
P3C2A_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 56) | NC score | 0.442246 (rank : 65) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P3C2A_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 57) | NC score | 0.440654 (rank : 66) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 58) | NC score | 0.592815 (rank : 50) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
RASL1_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 59) | NC score | 0.579636 (rank : 52) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | O95294, Q52M03, Q96CC7 | Gene names | RASAL1, RASAL | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
RASL1_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 60) | NC score | 0.556681 (rank : 56) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
FA62A_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 61) | NC score | 0.699598 (rank : 47) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
FA62A_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 62) | NC score | 0.701428 (rank : 46) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
NED4L_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 63) | NC score | 0.395353 (rank : 77) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NEDD4_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 64) | NC score | 0.358497 (rank : 86) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
BAIP3_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 65) | NC score | 0.568676 (rank : 55) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O94812, O94839, Q2M226, Q658J2, Q76N05, Q96RZ3, Q9UJK1 | Gene names | BAIAP3, KIAA0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 66) | NC score | 0.367919 (rank : 84) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
NED4L_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 67) | NC score | 0.388601 (rank : 81) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
PA24D_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 68) | NC score | 0.438751 (rank : 67) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q50L43, Q3TJC5 | Gene names | Pla2g4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
BAIP3_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 69) | NC score | 0.569770 (rank : 54) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q80TT2, Q6RUT4 | Gene names | Baiap3, Gm937, Kiaa0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
SYT13_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 70) | NC score | 0.732879 (rank : 44) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q7L8C5, Q9BQS3, Q9H041, Q9P2C0 | Gene names | SYT13, KIAA1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
PA24E_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 71) | NC score | 0.329550 (rank : 90) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q50L42, Q8BX44 | Gene names | Pla2g4e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 epsilon (EC 3.1.1.4) (cPLA2-epsilon) (Phospholipase A2 group IVE). | |||||
|
DYSF_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 72) | NC score | 0.428797 (rank : 69) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q9ESD7, Q9QXC0 | Gene names | Dysf, Fer1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
MYOF_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 73) | NC score | 0.444391 (rank : 64) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q9NZM1, Q9HBU3, Q9NZM0, Q9ULL3, Q9Y4U4 | Gene names | FER1L3, KIAA1207, MYOF | |||
|
Domain Architecture |

|
|||||
| Description | Myoferlin (Fer-1-like protein 3). | |||||
|
DYSF_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 74) | NC score | 0.431437 (rank : 68) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | O75923, O75696, Q9UEN7 | Gene names | DYSF, FER1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 75) | NC score | 0.192005 (rank : 119) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
PA24E_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 76) | NC score | 0.307688 (rank : 91) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q3MJ16, Q6ZSC0 | Gene names | PLA2G4E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 epsilon (EC 3.1.1.4) (cPLA2-epsilon) (Phospholipase A2 group IVE). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 77) | NC score | 0.206144 (rank : 117) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
PA24D_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 78) | NC score | 0.388892 (rank : 80) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q86XP0, Q8N176 | Gene names | PLA2G4D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
CPNE2_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 79) | NC score | 0.280581 (rank : 98) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P59108, Q8K1D7 | Gene names | Cpne2 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-2 (Copine II). | |||||
|
SYT13_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 80) | NC score | 0.727981 (rank : 45) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9EQT6, Q6ZPR2, Q8BRK6 | Gene names | Syt13, Kiaa1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
CPNE2_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 81) | NC score | 0.276596 (rank : 100) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96FN4, Q68D19, Q86XP9 | Gene names | CPNE2 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-2 (Copine II). | |||||
|
SMUF1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 82) | NC score | 0.178164 (rank : 122) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
UN13B_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 83) | NC score | 0.396174 (rank : 76) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
K0528_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 84) | NC score | 0.502734 (rank : 59) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q86YS7, O60280, Q7Z619, Q86SU3 | Gene names | KIAA0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
K0528_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 85) | NC score | 0.501992 (rank : 60) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q7TPS5, Q3TQY6, Q6A052, Q80VA1, Q8C0U3, Q8CID5, Q9CS85 | Gene names | Kiaa0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
OTOF_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 86) | NC score | 0.390923 (rank : 79) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9HC10, Q9HC08, Q9HC09, Q9Y650 | Gene names | OTOF, FER1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
SMUF1_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 87) | NC score | 0.176293 (rank : 123) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
UN13A_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 88) | NC score | 0.394765 (rank : 78) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
UN13D_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 89) | NC score | 0.579311 (rank : 53) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q70J99, Q9H7K5 | Gene names | UNC13D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog D (Munc13-4). | |||||
|
OTOF_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 90) | NC score | 0.384068 (rank : 82) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9ESF1, Q9ESF2 | Gene names | Otof, Fer1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
UN13B_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 91) | NC score | 0.396243 (rank : 75) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 92) | NC score | 0.152337 (rank : 128) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 93) | NC score | 0.153352 (rank : 127) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
PA24B_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 94) | NC score | 0.348478 (rank : 89) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q4QQM1, Q80VV8, Q91W88 | Gene names | Pla2g4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 95) | NC score | 0.223390 (rank : 113) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
CPNE5_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 96) | NC score | 0.287528 (rank : 95) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8JZW4 | Gene names | Cpne5 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-5 (Copine V). | |||||
|
RIMS3_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 97) | NC score | 0.417267 (rank : 72) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UJD0, Q92511 | Gene names | RIMS3, KIAA0237 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
CPNE5_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 98) | NC score | 0.286485 (rank : 96) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9HCH3 | Gene names | CPNE5, KIAA1599 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-5 (Copine V). | |||||
|
RASA2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 99) | NC score | 0.426148 (rank : 70) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RIMS3_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 100) | NC score | 0.407339 (rank : 74) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q80U57 | Gene names | Rims3, Kiaa0237 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
CPNE3_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 101) | NC score | 0.300691 (rank : 93) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8BT60, Q5CZX9, Q5DU22 | Gene names | Cpne3, Kiaa0636 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-3 (Copine III). | |||||
|
RASA3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 102) | NC score | 0.423016 (rank : 71) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
RASA2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 103) | NC score | 0.414260 (rank : 73) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
CPNE8_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 104) | NC score | 0.277033 (rank : 99) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q86YQ8, Q2TB41 | Gene names | CPNE8 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
CPNE3_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 105) | NC score | 0.301893 (rank : 92) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O75131, Q8IYA1 | Gene names | CPNE3, CPN3, KIAA0636 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-3 (Copine III). | |||||
|
CPNE8_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 106) | NC score | 0.271408 (rank : 103) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9DC53, Q9CUZ8 | Gene names | Cpne8 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
SMUF2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 107) | NC score | 0.170914 (rank : 124) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
RIMS4_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 108) | NC score | 0.482222 (rank : 62) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P60191 | Gene names | Rims4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
PLCB1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 109) | NC score | 0.047945 (rank : 171) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
RASA3_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 110) | NC score | 0.355868 (rank : 88) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
RIMS4_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 111) | NC score | 0.479258 (rank : 63) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9H426, Q5JWT7 | Gene names | RIMS4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
PA24B_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 112) | NC score | 0.286210 (rank : 97) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O95712, Q59GF9, Q8TB10, Q9UKV7 | Gene names | PLA2G4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
PA24A_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 113) | NC score | 0.293753 (rank : 94) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P47713 | Gene names | Pla2g4a, Cpla2, Pla2g4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
PA24F_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 114) | NC score | 0.244275 (rank : 104) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q50L41, Q80VQ8, Q80VV9 | Gene names | Pla2g4f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 zeta (EC 3.1.1.4) (cPLA2-zeta) (Phospholipase A2 group IVF). | |||||
|
PLCB2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 115) | NC score | 0.046850 (rank : 172) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
RFIP2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 116) | NC score | 0.372001 (rank : 83) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q7L804, Q9Y2F0 | Gene names | RAB11FIP2, KIAA0941 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 2 (Rab11-FIP2) (NRip11). | |||||
|
PLCD1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 117) | NC score | 0.206067 (rank : 118) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P51178 | Gene names | PLCD1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
CPNE7_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 118) | NC score | 0.237905 (rank : 108) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UBL6 | Gene names | CPNE7 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-7 (Copine VII). | |||||
|
PLCG2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 119) | NC score | 0.058218 (rank : 151) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16885, Q969T5 | Gene names | PLCG2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-2) (Phospholipase C-gamma-2) (PLC-IV). | |||||
|
CPNE6_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 120) | NC score | 0.214089 (rank : 116) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Z140 | Gene names | Cpne6 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-6 (Copine VI) (Neuronal-copine) (N-copine). | |||||
|
PLCB1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 121) | NC score | 0.043410 (rank : 175) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
PA24A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 122) | NC score | 0.274671 (rank : 102) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P47712 | Gene names | PLA2G4A, CPLA2, PLA2G4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
KPCE_HUMAN
|
||||||
| θ value | 0.279714 (rank : 123) | NC score | 0.044025 (rank : 173) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02156, Q53SL4, Q53SM5, Q9UE81 | Gene names | PRKCE, PKCE | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C epsilon type (EC 2.7.11.13) (nPKC-epsilon). | |||||
|
KPCE_MOUSE
|
||||||
| θ value | 0.279714 (rank : 124) | NC score | 0.043737 (rank : 174) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P16054 | Gene names | Prkce, Pkce, Pkcea | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C epsilon type (EC 2.7.11.13) (nPKC-epsilon). | |||||
|
CPNE1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 125) | NC score | 0.275612 (rank : 101) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8C166, Q925K4, Q925K5 | Gene names | Cpne1 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-1 (Copine I). | |||||
|
PA24F_HUMAN
|
||||||
| θ value | 0.365318 (rank : 126) | NC score | 0.215789 (rank : 114) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q68DD2, Q6ZMC8 | Gene names | PLA2G4F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 zeta (EC 3.1.1.4) (cPLA2-zeta) (Phospholipase A2 group IVF). | |||||
|
WNK3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 127) | NC score | 0.020427 (rank : 178) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
CPNE6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 128) | NC score | 0.214890 (rank : 115) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O95741, Q53HA6, Q8WVG1 | Gene names | CPNE6 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-6 (Copine VI) (Neuronal-copine) (N-copine). | |||||
|
WWC1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 129) | NC score | 0.164742 (rank : 125) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 130) | NC score | 0.015960 (rank : 180) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_MOUSE
|
||||||
| θ value | 0.813845 (rank : 131) | NC score | 0.015970 (rank : 179) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
PLCD1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 132) | NC score | 0.149092 (rank : 130) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8R3B1 | Gene names | Plcd1, Plcd | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
PERF_MOUSE
|
||||||
| θ value | 1.81305 (rank : 133) | NC score | 0.056673 (rank : 155) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10820 | Gene names | Prf1, Pfp | |||
|
Domain Architecture |

|
|||||
| Description | Perforin-1 precursor (P1) (Lymphocyte pore-forming protein) (Cytolysin). | |||||
|
CLH2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 134) | NC score | 0.007498 (rank : 185) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
NDK7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 135) | NC score | 0.009241 (rank : 184) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5B8, Q5TGZ4 | Gene names | NME7 | |||
|
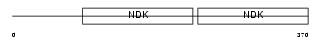
Domain Architecture |
|
|||||
| Description | Nucleoside diphosphate kinase 7 (EC 2.7.4.6) (NDK 7) (NDP kinase 7) (nm23-H7). | |||||
|
RASA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 136) | NC score | 0.078276 (rank : 140) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20936 | Gene names | RASA1, RASA | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 1 (GTPase-activating protein) (GAP) (Ras p21 protein activator) (p120GAP) (RasGAP). | |||||
|
CPNE4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 137) | NC score | 0.190504 (rank : 120) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96A23, Q8TEX1 | Gene names | CPNE4 | |||
|
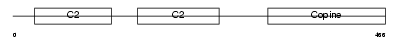
Domain Architecture |
|
|||||
| Description | Copine-4 (Copine IV) (Copine-8). | |||||
|
CPNE4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 138) | NC score | 0.188023 (rank : 121) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BLR2 | Gene names | Cpne4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Copine-4 (Copine IV). | |||||
|
NTAN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.015875 (rank : 181) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96AB6 | Gene names | NTAN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein N-terminal asparagine amidohydrolase (EC 3.5.1.-) (Protein NH2-terminal asparagine deamidase) (N-terminal Asn amidase) (NTN- amidase) (PNAD) (Protein NH2-terminal asparagine amidohydrolase) (PNAA). | |||||
|
RPGR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.012139 (rank : 182) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
CCD1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.039156 (rank : 177) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P1N0, Q7Z435, Q86XV0, Q8NF89, Q9H603, Q9NXI1 | Gene names | CC2D1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1) (Putative NF-kappa-B- activating protein 023N). | |||||
|
CCD1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.041581 (rank : 176) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K1A6, Q2MJB5, Q8R3Z4 | Gene names | Cc2d1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1). | |||||
|
NTAN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.010511 (rank : 183) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64311 | Gene names | Ntan1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein N-terminal asparagine amidohydrolase (EC 3.5.1.-) (Protein NH2-terminal asparagine deamidase) (N-terminal Asn amidase) (NTN- amidase) (PNAD) (Protein NH2-terminal asparagine amidohydrolase) (PNAA). | |||||
|
P3C2G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.134454 (rank : 132) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
BSN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.053714 (rank : 160) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.054138 (rank : 159) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CPNE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.239732 (rank : 107) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q99829, Q6IBL3, Q9H243, Q9NTZ7 | Gene names | CPNE1, CPN1 | |||
|
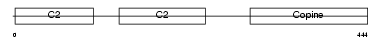
Domain Architecture |
|
|||||
| Description | Copine-1 (Copine I). | |||||
|
DAB2P_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.059845 (rank : 148) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VWQ8, Q8TDL2, Q96SE1, Q9C0C0 | Gene names | DAB2IP, AF9Q34, AIP1, KIAA1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein) (DAB2 interaction protein) (ASK-interacting protein 1). | |||||
|
DAB2P_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.057694 (rank : 152) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UHC7, Q3TPD5, Q3UH44, Q6JTV1, Q80T97 | Gene names | Dab2ip, Kiaa1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein). | |||||
|
DGKQ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.052338 (rank : 162) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P52824 | Gene names | DGKQ, DAGK4 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase theta (EC 2.7.1.107) (Diglyceride kinase theta) (DGK-theta) (DAG kinase theta). | |||||
|
GRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.058842 (rank : 149) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IV61, O94931 | Gene names | RASGRP3, GRP3, KIAA0846 | |||
|
Domain Architecture |

|
|||||
| Description | RAS guanyl-releasing protein 3 (Calcium and DAG-regulated guanine nucleotide exchange factor III) (Guanine nucleotide exchange factor for Rap1). | |||||
|
HECD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.052102 (rank : 165) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.052840 (rank : 161) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.056130 (rank : 157) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.056489 (rank : 156) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
ITCH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.068789 (rank : 143) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
ITCH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.066144 (rank : 144) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
K0317_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.057445 (rank : 154) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.057532 (rank : 153) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
MELPH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.113938 (rank : 136) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
MELPH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.113934 (rank : 137) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.129039 (rank : 134) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
MYRIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.133702 (rank : 133) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
P3C2G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.163829 (rank : 126) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PA24C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.084361 (rank : 139) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UP65, O75457, Q9UG68 | Gene names | PLA2G4C | |||
|
Domain Architecture |
|
|||||
| Description | Cytosolic phospholipase A2 gamma precursor (EC 3.1.1.4) (cPLA2-gamma) (Phospholipase A2 group IVC). | |||||
|
PERF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.058481 (rank : 150) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P14222, Q86WX7 | Gene names | PRF1, PFP | |||
|
Domain Architecture |

|
|||||
| Description | Perforin-1 precursor (P1) (Lymphocyte pore-forming protein) (PFP) (Cytolysin). | |||||
|
PLCG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.050804 (rank : 168) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19174, Q2V575 | Gene names | PLCG1, PLC1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
RASF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.050707 (rank : 169) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99MK9, Q9WUF5 | Gene names | Rassf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing protein 1 (Protein 123F2). | |||||
|
RFIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.141212 (rank : 131) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
RFIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.151279 (rank : 129) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
SLC2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.060592 (rank : 146) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NEV8, Q9Y4D6 | Gene names | EXPH5, KIAA0624, SLAC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Slp homolog lacking C2 domains b (Exophilin-5). | |||||
|
SYGP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.050272 (rank : 170) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
SYT8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.102413 (rank : 138) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NBV8, Q9NSV9 | Gene names | SYT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-8 (Synaptotagmin VIII) (SytVIII). | |||||
|
TAC2N_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.359386 (rank : 85) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N9U0 | Gene names | MTAC2D1, C14orf47 | |||
|
Domain Architecture |
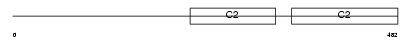
|
|||||
| Description | Membrane targeting tandem C2 domain-containing protein 1 (Tandem C2 protein in nucleus) (Tac2-N). | |||||
|
TAC2N_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.357957 (rank : 87) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91XT6, Q9D4A0 | Gene names | Mtac2d1 | |||
|
Domain Architecture |
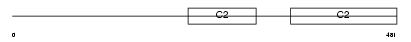
|
|||||
| Description | Membrane targeting tandem C2 domain-containing protein 1 (Tandem C2 protein in nucleus) (Tac2-N). | |||||
|
TOLIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.055828 (rank : 158) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QZ06, Q543N1, Q9D5P5 | Gene names | Tollip | |||
|
Domain Architecture |
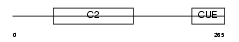
|
|||||
| Description | Toll-interacting protein. | |||||
|
UBE3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.051606 (rank : 166) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.051099 (rank : 167) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
UBE3C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.052131 (rank : 163) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.052110 (rank : 164) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
WWC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.116622 (rank : 135) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.070189 (rank : 141) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
WWP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.069665 (rank : 142) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
WWP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.060490 (rank : 147) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
WWP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.061900 (rank : 145) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
DOC2A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.97187e-173 (rank : 1) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
|
DOC2A_MOUSE
|
||||||
| NC score | 0.998683 (rank : 2) | θ value | 1.15563e-172 (rank : 2) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q7TNF0 | Gene names | Doc2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha). | |||||
|
DOC2B_HUMAN
|
||||||
| NC score | 0.989682 (rank : 3) | θ value | 1.48236e-119 (rank : 3) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q14184 | Gene names | DOC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
DOC2B_MOUSE
|
||||||
| NC score | 0.988803 (rank : 4) | θ value | 2.36663e-117 (rank : 4) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | P70169, Q6NXK3 | Gene names | Doc2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
DOC2G_MOUSE
|
||||||
| NC score | 0.974548 (rank : 5) | θ value | 9.69798e-87 (rank : 7) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9ESN1, Q6P8R8 | Gene names | Doc2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein gamma (Doc2-gamma). | |||||
|
RP3A_HUMAN
|
||||||
| NC score | 0.966335 (rank : 6) | θ value | 5.28454e-109 (rank : 5) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.955150 (rank : 7) | θ value | 1.75921e-104 (rank : 6) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SYT7_MOUSE
|
||||||
| NC score | 0.928090 (rank : 8) | θ value | 5.0505e-35 (rank : 11) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q9R0N7 | Gene names | Syt7 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
SYT7_HUMAN
|
||||||
| NC score | 0.926111 (rank : 9) | θ value | 2.86786e-30 (rank : 22) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | O43581 | Gene names | SYT7 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
SYT1_HUMAN
|
||||||
| NC score | 0.918293 (rank : 10) | θ value | 8.61488e-35 (rank : 12) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | P21579 | Gene names | SYT1, SYT | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT1_MOUSE
|
||||||
| NC score | 0.918271 (rank : 11) | θ value | 8.61488e-35 (rank : 13) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | P46096 | Gene names | Syt1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT2_MOUSE
|
||||||
| NC score | 0.918232 (rank : 12) | θ value | 1.01765e-35 (rank : 9) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | P46097, Q8R0E1 | Gene names | Syt2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT2_HUMAN
|
||||||
| NC score | 0.918155 (rank : 13) | θ value | 1.73584e-35 (rank : 10) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | Q8N9I0, Q496K5, Q8NBE5 | Gene names | SYT2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT5_HUMAN
|
||||||
| NC score | 0.913803 (rank : 14) | θ value | 4.42448e-31 (rank : 21) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | O00445, Q86X72 | Gene names | SYT5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV). | |||||
|
SYT5_MOUSE
|
||||||
| NC score | 0.912346 (rank : 15) | θ value | 6.18819e-25 (rank : 29) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | Q9R0N5 | Gene names | Syt5, Syt9 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV) (Synaptotagmin IX). | |||||
|
SYT3_MOUSE
|
||||||
| NC score | 0.912204 (rank : 16) | θ value | 7.29293e-34 (rank : 17) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT3_HUMAN
|
||||||
| NC score | 0.912066 (rank : 17) | θ value | 8.61488e-35 (rank : 14) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT4_HUMAN
|
||||||
| NC score | 0.911245 (rank : 18) | θ value | 4.568e-36 (rank : 8) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9H2B2, Q9P2K4 | Gene names | SYT4, KIAA1342 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
SYT6_MOUSE
|
||||||
| NC score | 0.908291 (rank : 19) | θ value | 2.50655e-34 (rank : 16) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | Q9R0N8 | Gene names | Syt6 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
SYT4_MOUSE
|
||||||
| NC score | 0.907769 (rank : 20) | θ value | 2.50655e-34 (rank : 15) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | P40749, Q3UFC1, Q8BGH3, Q8BRL6 | Gene names | Syt4, Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
SYT6_HUMAN
|
||||||
| NC score | 0.906286 (rank : 21) | θ value | 8.06329e-33 (rank : 18) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | Q5T7P8 | Gene names | SYT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
SYT10_HUMAN
|
||||||
| NC score | 0.904711 (rank : 22) | θ value | 8.3442e-30 (rank : 23) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q6XYQ8 | Gene names | SYT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
SYT10_MOUSE
|
||||||
| NC score | 0.903191 (rank : 23) | θ value | 4.14116e-29 (rank : 24) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q9R0N4 | Gene names | Syt10 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
SYT11_MOUSE
|
||||||
| NC score | 0.901513 (rank : 24) | θ value | 3.06405e-32 (rank : 20) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q9R0N3 | Gene names | Syt11 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
SYT11_HUMAN
|
||||||
| NC score | 0.900937 (rank : 25) | θ value | 3.06405e-32 (rank : 19) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9BT88, Q14998, Q68CT5, Q8IXU3, Q96SU2 | Gene names | SYT11, KIAA0080 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
SYT9_HUMAN
|
||||||
| NC score | 0.895939 (rank : 26) | θ value | 2.96777e-27 (rank : 26) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q86SS6 | Gene names | SYT9 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX). | |||||
|
SYT9_MOUSE
|
||||||
| NC score | 0.895578 (rank : 27) | θ value | 8.63488e-27 (rank : 28) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | Q9R0N9, Q8C280 | Gene names | Syt9, Syt5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX) (Synaptotagmin V). | |||||
|
SYT12_MOUSE
|
||||||
| NC score | 0.855576 (rank : 28) | θ value | 3.51386e-20 (rank : 32) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q920N7 | Gene names | Syt12 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYTL4_HUMAN
|
||||||
| NC score | 0.854962 (rank : 29) | θ value | 5.97985e-28 (rank : 25) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYT15_HUMAN
|
||||||
| NC score | 0.854137 (rank : 30) | θ value | 1.5773e-20 (rank : 31) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9BQS2, Q7Z439, Q7Z440 | Gene names | SYT15 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV) (Chr10Syt). | |||||
|
SYT12_HUMAN
|
||||||
| NC score | 0.853177 (rank : 31) | θ value | 3.88503e-19 (rank : 33) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q8IV01 | Gene names | SYT12 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYTL4_MOUSE
|
||||||
| NC score | 0.849046 (rank : 32) | θ value | 6.61148e-27 (rank : 27) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9R0Q1, Q8R321, Q9R0Q0 | Gene names | Sytl4, Slp4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYT8_MOUSE
|
||||||
| NC score | 0.837063 (rank : 33) | θ value | 1.42992e-13 (rank : 52) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9R0N6, Q64366, Q8C6E7 | Gene names | Syt8 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-8 (Synaptotagmin VIII) (SytVIII). | |||||
|
SYT15_MOUSE
|
||||||
| NC score | 0.830647 (rank : 34) | θ value | 1.13037e-18 (rank : 34) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8C6N3, Q7TN81, Q7TN82, Q8C999 | Gene names | Syt15 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV). | |||||
|
SYTL5_HUMAN
|
||||||
| NC score | 0.820143 (rank : 35) | θ value | 1.99067e-23 (rank : 30) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
SYTL1_HUMAN
|
||||||
| NC score | 0.811403 (rank : 36) | θ value | 1.24977e-17 (rank : 35) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
SYTL5_MOUSE
|
||||||
| NC score | 0.810765 (rank : 37) | θ value | 3.63628e-17 (rank : 36) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.796487 (rank : 38) | θ value | 6.85773e-16 (rank : 39) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
SYTL2_MOUSE
|
||||||
| NC score | 0.791535 (rank : 39) | θ value | 1.99529e-15 (rank : 43) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
SYTL3_MOUSE
|
||||||
| NC score | 0.781794 (rank : 40) | θ value | 5.25075e-16 (rank : 38) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.777139 (rank : 41) | θ value | 4.91457e-14 (rank : 49) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
SYT14_MOUSE
|
||||||
| NC score | 0.758356 (rank : 42) | θ value | 1.52774e-15 (rank : 41) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7TN84 | Gene names | Syt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
SYT14_HUMAN
|
||||||
| NC score | 0.753649 (rank : 43) | θ value | 3.76295e-14 (rank : 48) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NB59, Q5THX7, Q707N3, Q707N4, Q707N5, Q707N6, Q707N7 | Gene names | SYT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
SYT13_HUMAN
|
||||||
| NC score | 0.732879 (rank : 44) | θ value | 1.69304e-06 (rank : 70) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q7L8C5, Q9BQS3, Q9H041, Q9P2C0 | Gene names | SYT13, KIAA1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
SYT13_MOUSE
|
||||||
| NC score | 0.727981 (rank : 45) | θ value | 0.000158464 (rank : 80) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9EQT6, Q6ZPR2, Q8BRK6 | Gene names | Syt13, Kiaa1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
FA62A_MOUSE
|
||||||
| NC score | 0.701428 (rank : 46) | θ value | 1.38499e-08 (rank : 62) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
FA62A_HUMAN
|
||||||
| NC score | 0.699598 (rank : 47) | θ value | 6.21693e-09 (rank : 61) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
RASL2_HUMAN
|
||||||
| NC score | 0.672344 (rank : 48) | θ value | 7.58209e-15 (rank : 46) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | O43374, O60286, Q86UW3, Q96QU0 | Gene names | RASA4, CAPRI, GAPL, KIAA0538 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2) (Calcium-promoted Ras inactivator). | |||||
|
RIMS2_HUMAN
|
||||||
| NC score | 0.597378 (rank : 49) | θ value | 2.28291e-11 (rank : 54) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.592815 (rank : 50) | θ value | 6.64225e-11 (rank : 58) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
RIMS2_MOUSE
|
||||||
| NC score | 0.590733 (rank : 51) | θ value | 2.28291e-11 (rank : 55) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
RASL1_HUMAN
|
||||||
| NC score | 0.579636 (rank : 52) | θ value | 8.67504e-11 (rank : 59) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | O95294, Q52M03, Q96CC7 | Gene names | RASAL1, RASAL | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
UN13D_HUMAN
|
||||||
| NC score | 0.579311 (rank : 53) | θ value | 0.000270298 (rank : 89) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q70J99, Q9H7K5 | Gene names | UNC13D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog D (Munc13-4). | |||||
|
BAIP3_MOUSE
|
||||||
| NC score | 0.569770 (rank : 54) | θ value | 9.92553e-07 (rank : 69) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q80TT2, Q6RUT4 | Gene names | Baiap3, Gm937, Kiaa0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
BAIP3_HUMAN
|
||||||
| NC score | 0.568676 (rank : 55) | θ value | 3.41135e-07 (rank : 65) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O94812, O94839, Q2M226, Q658J2, Q76N05, Q96RZ3, Q9UJK1 | Gene names | BAIAP3, KIAA0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
RASL1_MOUSE
|
||||||
| NC score | 0.556681 (rank : 56) | θ value | 1.25267e-09 (rank : 60) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.546740 (rank : 57) | θ value | 1.058e-16 (rank : 37) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.516303 (rank : 58) | θ value | 1.16975e-15 (rank : 40) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
K0528_HUMAN
|
||||||
| NC score | 0.502734 (rank : 59) | θ value | 0.000270298 (rank : 84) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q86YS7, O60280, Q7Z619, Q86SU3 | Gene names | KIAA0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
K0528_MOUSE
|
||||||
| NC score | 0.501992 (rank : 60) | θ value | 0.000270298 (rank : 85) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q7TPS5, Q3TQY6, Q6A052, Q80VA1, Q8C0U3, Q8CID5, Q9CS85 | Gene names | Kiaa0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
P3C2B_HUMAN
|
||||||
| NC score | 0.499957 (rank : 61) | θ value | 1.42992e-13 (rank : 51) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
RIMS4_MOUSE
|
||||||
| NC score | 0.482222 (rank : 62) | θ value | 0.00869519 (rank : 108) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P60191 | Gene names | Rims4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
RIMS4_HUMAN
|
||||||
| NC score | 0.479258 (rank : 63) | θ value | 0.0113563 (rank : 111) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9H426, Q5JWT7 | Gene names | RIMS4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
MYOF_HUMAN
|
||||||
| NC score | 0.444391 (rank : 64) | θ value | 2.44474e-05 (rank : 73) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q9NZM1, Q9HBU3, Q9NZM0, Q9ULL3, Q9Y4U4 | Gene names | FER1L3, KIAA1207, MYOF | |||
|
Domain Architecture |

|
|||||
| Description | Myoferlin (Fer-1-like protein 3). | |||||
|
P3C2A_HUMAN
|
||||||
| NC score | 0.442246 (rank : 65) | θ value | 6.64225e-11 (rank : 56) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P3C2A_MOUSE
|
||||||
| NC score | 0.440654 (rank : 66) | θ value | 6.64225e-11 (rank : 57) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
PA24D_MOUSE
|
||||||
| NC score | 0.438751 (rank : 67) | θ value | 7.59969e-07 (rank : 68) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q50L43, Q3TJC5 | Gene names | Pla2g4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
DYSF_HUMAN
|
||||||
| NC score | 0.431437 (rank : 68) | θ value | 3.19293e-05 (rank : 74) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | O75923, O75696, Q9UEN7 | Gene names | DYSF, FER1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
DYSF_MOUSE
|
||||||
| NC score | 0.428797 (rank : 69) | θ value | 8.40245e-06 (rank : 72) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q9ESD7, Q9QXC0 | Gene names | Dysf, Fer1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
RASA2_MOUSE
|
||||||
| NC score | 0.426148 (rank : 70) | θ value | 0.00175202 (rank : 99) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA3_HUMAN
|
||||||
| NC score | 0.423016 (rank : 71) | θ value | 0.00228821 (rank : 102) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
RIMS3_HUMAN
|
||||||
| NC score | 0.417267 (rank : 72) | θ value | 0.00134147 (rank : 97) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UJD0, Q92511 | Gene names | RIMS3, KIAA0237 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
RASA2_HUMAN
|
||||||
| NC score | 0.414260 (rank : 73) | θ value | 0.00390308 (rank : 103) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RIMS3_MOUSE
|
||||||
| NC score | 0.407339 (rank : 74) | θ value | 0.00175202 (rank : 100) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q80U57 | Gene names | Rims3, Kiaa0237 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
UN13B_MOUSE
|
||||||
| NC score | 0.396243 (rank : 75) | θ value | 0.00035302 (rank : 91) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
UN13B_HUMAN
|
||||||
| NC score | 0.396174 (rank : 76) | θ value | 0.00020696 (rank : 83) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
NED4L_HUMAN
|
||||||
| NC score | 0.395353 (rank : 77) | θ value | 5.26297e-08 (rank : 63) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
UN13A_HUMAN
|
||||||
| NC score | 0.394765 (rank : 78) | θ value | 0.000270298 (rank : 88) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
OTOF_HUMAN
|
||||||
| NC score | 0.390923 (rank : 79) | θ value | 0.000270298 (rank : 86) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9HC10, Q9HC08, Q9HC09, Q9Y650 | Gene names | OTOF, FER1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
PA24D_HUMAN
|
||||||
| NC score | 0.388892 (rank : 80) | θ value | 9.29e-05 (rank : 78) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q86XP0, Q8N176 | Gene names | PLA2G4D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
NED4L_MOUSE
|
||||||
| NC score | 0.388601 (rank : 81) | θ value | 5.81887e-07 (rank : 67) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
OTOF_MOUSE
|
||||||
| NC score | 0.384068 (rank : 82) | θ value | 0.00035302 (rank : 90) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9ESF1, Q9ESF2 | Gene names | Otof, Fer1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
RFIP2_HUMAN
|
||||||
| NC score | 0.372001 (rank : 83) | θ value | 0.0431538 (rank : 116) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q7L804, Q9Y2F0 | Gene names | RAB11FIP2, KIAA0941 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 2 (Rab11-FIP2) (NRip11). | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.367919 (rank : 84) | θ value | 4.45536e-07 (rank : 66) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
TAC2N_HUMAN
|
||||||
| NC score | 0.359386 (rank : 85) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N9U0 | Gene names | MTAC2D1, C14orf47 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane targeting tandem C2 domain-containing protein 1 (Tandem C2 protein in nucleus) (Tac2-N). | |||||
|
NEDD4_MOUSE
|
||||||
| NC score | 0.358497 (rank : 86) | θ value | 5.26297e-08 (rank : 64) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
TAC2N_MOUSE
|
||||||
| NC score | 0.357957 (rank : 87) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91XT6, Q9D4A0 | Gene names | Mtac2d1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane targeting tandem C2 domain-containing protein 1 (Tandem C2 protein in nucleus) (Tac2-N). | |||||
|
RASA3_MOUSE
|
||||||
| NC score | 0.355868 (rank : 88) | θ value | 0.0113563 (rank : 110) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
PA24B_MOUSE
|
||||||
| NC score | 0.348478 (rank : 89) | θ value | 0.000786445 (rank : 94) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q4QQM1, Q80VV8, Q91W88 | Gene names | Pla2g4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
PA24E_MOUSE
|
||||||
| NC score | 0.329550 (rank : 90) | θ value | 6.43352e-06 (rank : 71) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q50L42, Q8BX44 | Gene names | Pla2g4e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 epsilon (EC 3.1.1.4) (cPLA2-epsilon) (Phospholipase A2 group IVE). | |||||
|
PA24E_HUMAN
|
||||||
| NC score | 0.307688 (rank : 91) | θ value | 7.1131e-05 (rank : 76) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q3MJ16, Q6ZSC0 | Gene names | PLA2G4E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 epsilon (EC 3.1.1.4) (cPLA2-epsilon) (Phospholipase A2 group IVE). | |||||
|
CPNE3_HUMAN
|
||||||
| NC score | 0.301893 (rank : 92) | θ value | 0.00665767 (rank : 105) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O75131, Q8IYA1 | Gene names | CPNE3, CPN3, KIAA0636 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-3 (Copine III). | |||||
|
CPNE3_MOUSE
|
||||||
| NC score | 0.300691 (rank : 93) | θ value | 0.00228821 (rank : 101) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8BT60, Q5CZX9, Q5DU22 | Gene names | Cpne3, Kiaa0636 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-3 (Copine III). | |||||
|
PA24A_MOUSE
|
||||||
| NC score | 0.293753 (rank : 94) | θ value | 0.0252991 (rank : 113) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P47713 | Gene names | Pla2g4a, Cpla2, Pla2g4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
CPNE5_MOUSE
|
||||||
| NC score | 0.287528 (rank : 95) | θ value | 0.00134147 (rank : 96) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8JZW4 | Gene names | Cpne5 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-5 (Copine V). | |||||
|
CPNE5_HUMAN
|
||||||
| NC score | 0.286485 (rank : 96) | θ value | 0.00175202 (rank : 98) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9HCH3 | Gene names | CPNE5, KIAA1599 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-5 (Copine V). | |||||
|
PA24B_HUMAN
|
||||||
| NC score | 0.286210 (rank : 97) | θ value | 0.0148317 (rank : 112) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O95712, Q59GF9, Q8TB10, Q9UKV7 | Gene names | PLA2G4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
CPNE2_MOUSE
|
||||||
| NC score | 0.280581 (rank : 98) | θ value | 0.000158464 (rank : 79) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P59108, Q8K1D7 | Gene names | Cpne2 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-2 (Copine II). | |||||
|
CPNE8_HUMAN
|
||||||
| NC score | 0.277033 (rank : 99) | θ value | 0.00509761 (rank : 104) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q86YQ8, Q2TB41 | Gene names | CPNE8 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
CPNE2_HUMAN
|
||||||
| NC score | 0.276596 (rank : 100) | θ value | 0.00020696 (rank : 81) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96FN4, Q68D19, Q86XP9 | Gene names | CPNE2 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-2 (Copine II). | |||||
|
CPNE1_MOUSE
|
||||||
| NC score | 0.275612 (rank : 101) | θ value | 0.365318 (rank : 125) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8C166, Q925K4, Q925K5 | Gene names | Cpne1 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-1 (Copine I). | |||||
|
PA24A_HUMAN
|
||||||
| NC score | 0.274671 (rank : 102) | θ value | 0.21417 (rank : 122) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P47712 | Gene names | PLA2G4A, CPLA2, PLA2G4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
CPNE8_MOUSE
|
||||||
| NC score | 0.271408 (rank : 103) | θ value | 0.00665767 (rank : 106) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9DC53, Q9CUZ8 | Gene names | Cpne8 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
PA24F_MOUSE
|
||||||
| NC score | 0.244275 (rank : 104) | θ value | 0.0431538 (rank : 114) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q50L41, Q80VQ8, Q80VV9 | Gene names | Pla2g4f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 zeta (EC 3.1.1.4) (cPLA2-zeta) (Phospholipase A2 group IVF). | |||||
|
KPCG_HUMAN
|
||||||
| NC score | 0.243175 (rank : 105) | θ value | 1.86753e-13 (rank : 53) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1126 | Shared Neighborhood Hits | 127 | |
| SwissProt Accessions | P05129 | Gene names | PRKCG, PKCG | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
KPCG_MOUSE
|
||||||
| NC score | 0.243081 (rank : 106) | θ value | 1.09485e-13 (rank : 50) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 127 | |
| SwissProt Accessions | P63318, P05697 | Gene names | Prkcg, Pkcc, Pkcg, Prkcc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
CPNE1_HUMAN
|
||||||
| NC score | 0.239732 (rank : 107) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q99829, Q6IBL3, Q9H243, Q9NTZ7 | Gene names | CPNE1, CPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-1 (Copine I). | |||||
|
CPNE7_HUMAN
|
||||||
| NC score | 0.237905 (rank : 108) | θ value | 0.0736092 (rank : 118) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UBL6 | Gene names | CPNE7 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-7 (Copine VII). | |||||
|
KPCA_HUMAN
|
||||||
| NC score | 0.228334 (rank : 109) | θ value | 4.44505e-15 (rank : 45) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | P17252, Q15137, Q96RE4 | Gene names | PRKCA, PKCA, PRKACA | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
KPCB_MOUSE
|
||||||
| NC score | 0.227823 (rank : 110) | θ value | 1.99529e-15 (rank : 42) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 127 | |
| SwissProt Accessions | P68404, P04410, P04411 | Gene names | Prkcb1, Pkcb, Prkcb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
KPCB_HUMAN
|
||||||
| NC score | 0.224895 (rank : 111) | θ value | 3.40345e-15 (rank : 44) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | P05771, O43744, P05127, Q15138, Q93060, Q9UE49, Q9UE50, Q9UEH8, Q9UJ30, Q9UJ33 | Gene names | PRKCB1, PKCB, PRKCB | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
KPCA_MOUSE
|
||||||
| NC score | 0.224336 (rank : 112) | θ value | 1.68911e-14 (rank : 47) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | P20444 | Gene names | Prkca, Pkca | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.223390 (rank : 113) | θ value | 0.000786445 (rank : 95) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
PA24F_HUMAN
|
||||||
| NC score | 0.215789 (rank : 114) | θ value | 0.365318 (rank : 126) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q68DD2, Q6ZMC8 | Gene names | PLA2G4F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 zeta (EC 3.1.1.4) (cPLA2-zeta) (Phospholipase A2 group IVF). | |||||
|
CPNE6_HUMAN
|
||||||
| NC score | 0.214890 (rank : 115) | θ value | 0.62314 (rank : 128) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O95741, Q53HA6, Q8WVG1 | Gene names | CPNE6 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-6 (Copine VI) (Neuronal-copine) (N-copine). | |||||
|
CPNE6_MOUSE
|
||||||
| NC score | 0.214089 (rank : 116) | θ value | 0.0961366 (rank : 120) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Z140 | Gene names | Cpne6 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-6 (Copine VI) (Neuronal-copine) (N-copine). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.206144 (rank : 117) | θ value | 9.29e-05 (rank : 77) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
PLCD1_HUMAN
|
||||||
| NC score | 0.206067 (rank : 118) | θ value | 0.0563607 (rank : 117) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P51178 | Gene names | PLCD1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.192005 (rank : 119) | θ value | 7.1131e-05 (rank : 75) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
CPNE4_HUMAN
|
||||||
| NC score | 0.190504 (rank : 120) | θ value | 4.03905 (rank : 137) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96A23, Q8TEX1 | Gene names | CPNE4 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-4 (Copine IV) (Copine-8). | |||||
|
CPNE4_MOUSE
|
||||||
| NC score | 0.188023 (rank : 121) | θ value | 4.03905 (rank : 138) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BLR2 | Gene names | Cpne4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Copine-4 (Copine IV). | |||||
|
SMUF1_HUMAN
|
||||||
| NC score | 0.178164 (rank : 122) | θ value | 0.00020696 (rank : 82) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF1_MOUSE
|
||||||
| NC score | 0.176293 (rank : 123) | θ value | 0.000270298 (rank : 87) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
SMUF2_HUMAN
|
||||||
| NC score | 0.170914 (rank : 124) | θ value | 0.00665767 (rank : 107) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
WWC1_MOUSE
|
||||||
| NC score | 0.164742 (rank : 125) | θ value | 0.62314 (rank : 129) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
P3C2G_MOUSE
|
||||||
| NC score | 0.163829 (rank : 126) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.153352 (rank : 127) | θ value | 0.000786445 (rank : 93) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.152337 (rank : 128) | θ value | 0.000786445 (rank : 92) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
RFIP1_MOUSE
|
||||||
| NC score | 0.151279 (rank : 129) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
PLCD1_MOUSE
|
||||||
| NC score | 0.149092 (rank : 130) | θ value | 1.06291 (rank : 132) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8R3B1 | Gene names | Plcd1, Plcd | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
RFIP1_HUMAN
|
||||||
| NC score | 0.141212 (rank : 131) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
P3C2G_HUMAN
|
||||||
| NC score | 0.134454 (rank : 132) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
MYRIP_MOUSE
|
||||||
| NC score | 0.133702 (rank : 133) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.129039 (rank : 134) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
WWC1_HUMAN
|
||||||
| NC score | 0.116622 (rank : 135) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
MELPH_HUMAN
|
||||||
| NC score | 0.113938 (rank : 136) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
MELPH_MOUSE
|
||||||
| NC score | 0.113934 (rank : 137) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
SYT8_HUMAN
|
||||||
| NC score | 0.102413 (rank : 138) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NBV8, Q9NSV9 | Gene names | SYT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-8 (Synaptotagmin VIII) (SytVIII). | |||||
|
PA24C_HUMAN
|
||||||
| NC score | 0.084361 (rank : 139) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UP65, O75457, Q9UG68 | Gene names | PLA2G4C | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 gamma precursor (EC 3.1.1.4) (cPLA2-gamma) (Phospholipase A2 group IVC). | |||||
|
RASA1_HUMAN
|
||||||
| NC score | 0.078276 (rank : 140) | θ value | 3.0926 (rank : 136) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20936 | Gene names | RASA1, RASA | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 1 (GTPase-activating protein) (GAP) (Ras p21 protein activator) (p120GAP) (RasGAP). | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.070189 (rank : 141) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
WWP1_MOUSE
|
||||||
| NC score | 0.069665 (rank : 142) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
ITCH_HUMAN
|
||||||
| NC score | 0.068789 (rank : 143) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
ITCH_MOUSE
|
||||||
| NC score | 0.066144 (rank : 144) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.061900 (rank : 145) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
SLC2B_HUMAN
|
||||||
| NC score | 0.060592 (rank : 146) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NEV8, Q9Y4D6 | Gene names | EXPH5, KIAA0624, SLAC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Slp homolog lacking C2 domains b (Exophilin-5). | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.060490 (rank : 147) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
DAB2P_HUMAN
|
||||||
| NC score | 0.059845 (rank : 148) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VWQ8, Q8TDL2, Q96SE1, Q9C0C0 | Gene names | DAB2IP, AF9Q34, AIP1, KIAA1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein) (DAB2 interaction protein) (ASK-interacting protein 1). | |||||
|
GRP3_HUMAN
|
||||||
| NC score | 0.058842 (rank : 149) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IV61, O94931 | Gene names | RASGRP3, GRP3, KIAA0846 | |||
|
Domain Architecture |

|
|||||
| Description | RAS guanyl-releasing protein 3 (Calcium and DAG-regulated guanine nucleotide exchange factor III) (Guanine nucleotide exchange factor for Rap1). | |||||
|
PERF_HUMAN
|
||||||
| NC score | 0.058481 (rank : 150) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P14222, Q86WX7 | Gene names | PRF1, PFP | |||
|
Domain Architecture |

|
|||||
| Description | Perforin-1 precursor (P1) (Lymphocyte pore-forming protein) (PFP) (Cytolysin). | |||||
|
PLCG2_HUMAN
|
||||||
| NC score | 0.058218 (rank : 151) | θ value | 0.0736092 (rank : 119) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16885, Q969T5 | Gene names | PLCG2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-2) (Phospholipase C-gamma-2) (PLC-IV). | |||||
|
DAB2P_MOUSE
|
||||||
| NC score | 0.057694 (rank : 152) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UHC7, Q3TPD5, Q3UH44, Q6JTV1, Q80T97 | Gene names | Dab2ip, Kiaa1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein). | |||||
|
K0317_MOUSE
|
||||||
| NC score | 0.057532 (rank : 153) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_HUMAN
|
||||||
| NC score | 0.057445 (rank : 154) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
PERF_MOUSE
|
||||||
| NC score | 0.056673 (rank : 155) | θ value | 1.81305 (rank : 133) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10820 | Gene names | Prf1, Pfp | |||
|
Domain Architecture |

|
|||||
| Description | Perforin-1 precursor (P1) (Lymphocyte pore-forming protein) (Cytolysin). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.056489 (rank : 156) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.056130 (rank : 157) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
TOLIP_MOUSE
|
||||||
| NC score | 0.055828 (rank : 158) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QZ06, Q543N1, Q9D5P5 | Gene names | Tollip | |||
|
Domain Architecture |

|
|||||
| Description | Toll-interacting protein. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.054138 (rank : 159) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.053714 (rank : 160) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
HECD2_MOUSE
|
||||||
| NC score | 0.052840 (rank : 161) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
DGKQ_HUMAN
|
||||||
| NC score | 0.052338 (rank : 162) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P52824 | Gene names | DGKQ, DAGK4 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase theta (EC 2.7.1.107) (Diglyceride kinase theta) (DGK-theta) (DAG kinase theta). | |||||
|
UBE3C_HUMAN
|
||||||
| NC score | 0.052131 (rank : 163) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| NC score | 0.052110 (rank : 164) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
HECD2_HUMAN
|
||||||
| NC score | 0.052102 (rank : 165) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
UBE3A_HUMAN
|
||||||
| NC score | 0.051606 (rank : 166) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3A_MOUSE
|
||||||
| NC score | 0.051099 (rank : 167) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
PLCG1_HUMAN
|
||||||
| NC score | 0.050804 (rank : 168) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19174, Q2V575 | Gene names | PLCG1, PLC1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
RASF1_MOUSE
|
||||||
| NC score | 0.050707 (rank : 169) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99MK9, Q9WUF5 | Gene names | Rassf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing protein 1 (Protein 123F2). | |||||
|
SYGP1_HUMAN
|
||||||
| NC score | 0.050272 (rank : 170) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
PLCB1_HUMAN
|
||||||
| NC score | 0.047945 (rank : 171) | θ value | 0.0113563 (rank : 109) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
PLCB2_HUMAN
|
||||||
| NC score | 0.046850 (rank : 172) | θ value | 0.0431538 (rank : 115) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
KPCE_HUMAN
|
||||||
| NC score | 0.044025 (rank : 173) | θ value | 0.279714 (rank : 123) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02156, Q53SL4, Q53SM5, Q9UE81 | Gene names | PRKCE, PKCE | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C epsilon type (EC 2.7.11.13) (nPKC-epsilon). | |||||
|
KPCE_MOUSE
|
||||||
| NC score | 0.043737 (rank : 174) | θ value | 0.279714 (rank : 124) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P16054 | Gene names | Prkce, Pkce, Pkcea | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C epsilon type (EC 2.7.11.13) (nPKC-epsilon). | |||||
|
PLCB1_MOUSE
|
||||||
| NC score | 0.043410 (rank : 175) | θ value | 0.125558 (rank : 121) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
CCD1A_MOUSE
|
||||||
| NC score | 0.041581 (rank : 176) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K1A6, Q2MJB5, Q8R3Z4 | Gene names | Cc2d1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1). | |||||
|
CCD1A_HUMAN
|
||||||
| NC score | 0.039156 (rank : 177) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P1N0, Q7Z435, Q86XV0, Q8NF89, Q9H603, Q9NXI1 | Gene names | CC2D1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1) (Putative NF-kappa-B- activating protein 023N). | |||||
|
WNK3_HUMAN
|
||||||
| NC score | 0.020427 (rank : 178) | θ value | 0.47712 (rank : 127) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
UBP5_MOUSE
|
||||||
| NC score | 0.015970 (rank : 179) | θ value | 0.813845 (rank : 131) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.015960 (rank : 180) | θ value | 0.813845 (rank : 130) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
NTAN1_HUMAN
|
||||||
| NC score | 0.015875 (rank : 181) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96AB6 | Gene names | NTAN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein N-terminal asparagine amidohydrolase (EC 3.5.1.-) (Protein NH2-terminal asparagine deamidase) (N-terminal Asn amidase) (NTN- amidase) (PNAD) (Protein NH2-terminal asparagine amidohydrolase) (PNAA). | |||||
|
RPGR1_HUMAN
|
||||||
| NC score | 0.012139 (rank : 182) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
NTAN1_MOUSE
|
||||||
| NC score | 0.010511 (rank : 183) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64311 | Gene names | Ntan1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein N-terminal asparagine amidohydrolase (EC 3.5.1.-) (Protein NH2-terminal asparagine deamidase) (N-terminal Asn amidase) (NTN- amidase) (PNAD) (Protein NH2-terminal asparagine amidohydrolase) (PNAA). | |||||
|
NDK7_HUMAN
|
||||||
| NC score | 0.009241 (rank : 184) | θ value | 3.0926 (rank : 135) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5B8, Q5TGZ4 | Gene names | NME7 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoside diphosphate kinase 7 (EC 2.7.4.6) (NDK 7) (NDP kinase 7) (nm23-H7). | |||||
|
CLH2_HUMAN
|
||||||
| NC score | 0.007498 (rank : 185) | θ value | 3.0926 (rank : 134) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||