Please be patient as the page loads
|
CLH2_HUMAN
|
||||||
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CLH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995191 (rank : 2) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q00610, Q6N0A0, Q86TF2 | Gene names | CLTC, CLH17, KIAA0034 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 1 (CLH-17). | |||||
|
CLH2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
CLH_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.995003 (rank : 3) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q68FD5 | Gene names | Cltc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin heavy chain. | |||||
|
VAV_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 4) | NC score | 0.022927 (rank : 15) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15498, Q15860 | Gene names | VAV1, VAV | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav. | |||||
|
VPS41_HUMAN
|
||||||
| θ value | 0.163984 (rank : 5) | NC score | 0.095128 (rank : 5) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49754, Q99851, Q99852 | Gene names | VPS41 | |||
|
Domain Architecture |
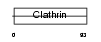
|
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (S53). | |||||
|
VPS41_MOUSE
|
||||||
| θ value | 0.163984 (rank : 6) | NC score | 0.095262 (rank : 4) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5KU39, Q3TPS1, Q80V99, Q8BKK6, Q8CG01, Q8CJC3 | Gene names | Vps41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (VAM2 homolog) (mVAM2). | |||||
|
WDR35_MOUSE
|
||||||
| θ value | 0.47712 (rank : 7) | NC score | 0.057483 (rank : 7) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BND3, Q8CDL4 | Gene names | Wdr35 | |||
|
Domain Architecture |
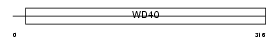
|
|||||
| Description | WD repeat protein 35. | |||||
|
ANLN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 8) | NC score | 0.026768 (rank : 8) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
KCC1D_HUMAN
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.004054 (rank : 38) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IU85, Q9HD31 | Gene names | CAMK1D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase type 1D (EC 2.7.11.17) (CaM kinase ID) (CaM kinase I delta) (CaMKI-delta) (CaM-KI delta) (CaMKI delta) (Camk1D) (CamKI-like protein kinase) (CKLiK). | |||||
|
KCC1D_MOUSE
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.004055 (rank : 37) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BW96, Q3U450, Q80W64, Q8BWI7 | Gene names | Camk1d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase type 1D (EC 2.7.11.17) (CaM kinase ID) (CaM kinase I delta) (CaMKI-delta) (CaM-KI delta) (CaMKI delta) (Camk1D) (CamKI-like protein kinase) (CKLiK) (mCKLiK). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.013504 (rank : 21) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.011660 (rank : 23) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
I2C2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 13) | NC score | 0.023399 (rank : 12) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKV8, Q8TCZ5, Q8WV58, Q96ID1 | Gene names | EIF2C2, AGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 2 (eIF2C 2) (eIF-2C 2) (Argonaute-2) (Slicer protein) (PAZ Piwi domain protein) (PPD). | |||||
|
I2C2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.023240 (rank : 13) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CJG0, Q4VAB3, Q571J6 | Gene names | Eif2c2, Ago2, Kiaa4215 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 2 (eIF2C 2) (eIF-2C 2) (Argonaute-2) (Slicer protein) (Piwi/argonaute family protein meIF2C2). | |||||
|
BSSP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.004932 (rank : 36) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZN4, O43342, Q6UXE0 | Gene names | PRSS22, BSSP4, PRSS26 | |||
|
Domain Architecture |
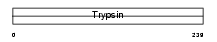
|
|||||
| Description | Brain-specific serine protease 4 precursor (EC 3.4.21.-) (BSSP-4) (Serine protease 22) (Tryptase epsilon). | |||||
|
DOC2A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.007498 (rank : 31) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
|
CSN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.025089 (rank : 10) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61201, O88950, Q15647, Q6FGP4, Q9BY54, Q9R249, Q9UNI2, Q9UNQ5 | Gene names | COPS2, CSN2, TRIP15 | |||
|
Domain Architecture |

|
|||||
| Description | COP9 signalosome complex subunit 2 (Signalosome subunit 2) (SGN2) (JAB1-containing signalosome subunit 2) (Thyroid receptor-interacting protein 15) (Alien homolog). | |||||
|
CSN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.025089 (rank : 11) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61202, O88950, Q15647, Q3V1W6, Q8R5B0, Q9CWU1, Q9R249, Q9UNQ5 | Gene names | Cops2, Csn2, Trip15 | |||
|
Domain Architecture |

|
|||||
| Description | COP9 signalosome complex subunit 2 (Signalosome subunit 2) (SGN2) (JAB1-containing signalosome subunit 2) (Thyroid receptor-interacting protein 15) (Alien homolog). | |||||
|
LRMP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.025162 (rank : 9) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60664, Q7TMU8 | Gene names | Lrmp, Jaw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphoid-restricted membrane protein (Protein Jaw1). | |||||
|
LRMP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.023093 (rank : 14) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12912, Q8N301 | Gene names | LRMP, JAW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphoid-restricted membrane protein (Protein Jaw1). | |||||
|
NRP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.016977 (rank : 16) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
UBP7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.009634 (rank : 28) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
1433B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.015954 (rank : 19) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31946 | Gene names | YWHAB | |||
|
Domain Architecture |
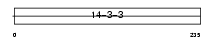
|
|||||
| Description | 14-3-3 protein beta/alpha (Protein kinase C inhibitor protein 1) (KCIP-1) (Protein 1054). | |||||
|
ABTB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.006962 (rank : 34) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969K4, Q6ZNU9, Q71MF1, Q96S62, Q96S63 | Gene names | ABTB1, BPOZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 1 (Elongation factor 1A-binding protein). | |||||
|
BSSP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.007706 (rank : 30) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ER10 | Gene names | Prss22, Bssp4, Prss26 | |||
|
Domain Architecture |
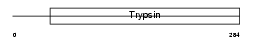
|
|||||
| Description | Brain-specific serine protease 4 precursor (EC 3.4.21.-) (BSSP-4) (Serine protease 22) (Tryptase epsilon). | |||||
|
CRNL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.011384 (rank : 26) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P63154, Q542E8, Q9CQC1 | Gene names | Crnkl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crooked neck-like protein 1 (Crooked neck homolog). | |||||
|
ZC11A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.011828 (rank : 22) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
DMAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.016429 (rank : 18) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPF5, Q5TG41, Q7Z3H7, Q9H0S8, Q9P2C2 | Gene names | DMAP1, KIAA1425 | |||
|
Domain Architecture |

|
|||||
| Description | DNA methyltransferase 1-associated protein 1 (DNMT1-associated protein 1) (DNMAP1). | |||||
|
DMAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.016447 (rank : 17) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JI44, Q99LM0, Q9CSS9, Q9DB33 | Gene names | Dmap1, Mmtr | |||
|
Domain Architecture |

|
|||||
| Description | DNA methyltransferase 1-associated protein 1 (DNMT1-associated protein 1) (DNMAP1) (MAT1-mediated transcriptional repressor). | |||||
|
DOC10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.006204 (rank : 35) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96BY6, O75178, Q9NW06, Q9NXI8 | Gene names | DOCK10, KIAA0694 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 10 (Protein zizimin 3). | |||||
|
K1HB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.001755 (rank : 39) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14525, O76010 | Gene names | KRTHA3B, HHA3-II, HKA3B | |||
|
Domain Architecture |
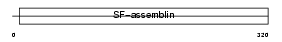
|
|||||
| Description | Keratin, type I cuticular Ha3-II (Hair keratin, type I Ha3-II). | |||||
|
NOC4L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.011532 (rank : 24) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHY2, Q8R1F7 | Gene names | Noc4l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 4 homolog (NOC4 protein homolog) (NOC4-like protein) (Nucleolar complex-associated protein 4-like protein). | |||||
|
NRP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.013805 (rank : 20) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.007181 (rank : 32) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
TAF1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.011422 (rank : 25) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZX4 | Gene names | TAF1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID 210 kDa subunit (TBP-associated factor 210 kDa) (TAF(II)210) (TBP-associated factor 1-like). | |||||
|
TAF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.011267 (rank : 27) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
TRAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.007744 (rank : 29) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12931, O43642, O75235, Q9UHL5 | Gene names | TRAP1, HSP75 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
WDR35_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.063374 (rank : 6) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2L0, Q8NE11 | Gene names | WDR35, KIAA1336 | |||
|
Domain Architecture |
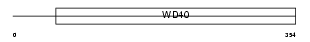
|
|||||
| Description | WD repeat protein 35. | |||||
|
ZFYV9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.007175 (rank : 33) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
CLH2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
CLH1_HUMAN
|
||||||
| NC score | 0.995191 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q00610, Q6N0A0, Q86TF2 | Gene names | CLTC, CLH17, KIAA0034 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 1 (CLH-17). | |||||
|
CLH_MOUSE
|
||||||
| NC score | 0.995003 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q68FD5 | Gene names | Cltc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin heavy chain. | |||||
|
VPS41_MOUSE
|
||||||
| NC score | 0.095262 (rank : 4) | θ value | 0.163984 (rank : 6) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5KU39, Q3TPS1, Q80V99, Q8BKK6, Q8CG01, Q8CJC3 | Gene names | Vps41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (VAM2 homolog) (mVAM2). | |||||
|
VPS41_HUMAN
|
||||||
| NC score | 0.095128 (rank : 5) | θ value | 0.163984 (rank : 5) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49754, Q99851, Q99852 | Gene names | VPS41 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (S53). | |||||
|
WDR35_HUMAN
|
||||||
| NC score | 0.063374 (rank : 6) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2L0, Q8NE11 | Gene names | WDR35, KIAA1336 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 35. | |||||
|
WDR35_MOUSE
|
||||||
| NC score | 0.057483 (rank : 7) | θ value | 0.47712 (rank : 7) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BND3, Q8CDL4 | Gene names | Wdr35 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 35. | |||||
|
ANLN_MOUSE
|
||||||
| NC score | 0.026768 (rank : 8) | θ value | 0.813845 (rank : 8) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
LRMP_MOUSE
|
||||||
| NC score | 0.025162 (rank : 9) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60664, Q7TMU8 | Gene names | Lrmp, Jaw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphoid-restricted membrane protein (Protein Jaw1). | |||||
|
CSN2_HUMAN
|
||||||
| NC score | 0.025089 (rank : 10) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61201, O88950, Q15647, Q6FGP4, Q9BY54, Q9R249, Q9UNI2, Q9UNQ5 | Gene names | COPS2, CSN2, TRIP15 | |||
|
Domain Architecture |

|
|||||
| Description | COP9 signalosome complex subunit 2 (Signalosome subunit 2) (SGN2) (JAB1-containing signalosome subunit 2) (Thyroid receptor-interacting protein 15) (Alien homolog). | |||||
|
CSN2_MOUSE
|
||||||
| NC score | 0.025089 (rank : 11) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61202, O88950, Q15647, Q3V1W6, Q8R5B0, Q9CWU1, Q9R249, Q9UNQ5 | Gene names | Cops2, Csn2, Trip15 | |||
|
Domain Architecture |

|
|||||
| Description | COP9 signalosome complex subunit 2 (Signalosome subunit 2) (SGN2) (JAB1-containing signalosome subunit 2) (Thyroid receptor-interacting protein 15) (Alien homolog). | |||||
|
I2C2_HUMAN
|
||||||
| NC score | 0.023399 (rank : 12) | θ value | 2.36792 (rank : 13) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKV8, Q8TCZ5, Q8WV58, Q96ID1 | Gene names | EIF2C2, AGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 2 (eIF2C 2) (eIF-2C 2) (Argonaute-2) (Slicer protein) (PAZ Piwi domain protein) (PPD). | |||||
|
I2C2_MOUSE
|
||||||
| NC score | 0.023240 (rank : 13) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CJG0, Q4VAB3, Q571J6 | Gene names | Eif2c2, Ago2, Kiaa4215 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 2 (eIF2C 2) (eIF-2C 2) (Argonaute-2) (Slicer protein) (Piwi/argonaute family protein meIF2C2). | |||||
|
LRMP_HUMAN
|
||||||
| NC score | 0.023093 (rank : 14) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12912, Q8N301 | Gene names | LRMP, JAW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphoid-restricted membrane protein (Protein Jaw1). | |||||
|
VAV_HUMAN
|
||||||
| NC score | 0.022927 (rank : 15) | θ value | 0.0193708 (rank : 4) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15498, Q15860 | Gene names | VAV1, VAV | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav. | |||||
|
NRP1_MOUSE
|
||||||
| NC score | 0.016977 (rank : 16) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
DMAP1_MOUSE
|
||||||
| NC score | 0.016447 (rank : 17) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JI44, Q99LM0, Q9CSS9, Q9DB33 | Gene names | Dmap1, Mmtr | |||
|
Domain Architecture |

|
|||||
| Description | DNA methyltransferase 1-associated protein 1 (DNMT1-associated protein 1) (DNMAP1) (MAT1-mediated transcriptional repressor). | |||||
|
DMAP1_HUMAN
|
||||||
| NC score | 0.016429 (rank : 18) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPF5, Q5TG41, Q7Z3H7, Q9H0S8, Q9P2C2 | Gene names | DMAP1, KIAA1425 | |||
|
Domain Architecture |

|
|||||
| Description | DNA methyltransferase 1-associated protein 1 (DNMT1-associated protein 1) (DNMAP1). | |||||
|
1433B_HUMAN
|
||||||
| NC score | 0.015954 (rank : 19) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31946 | Gene names | YWHAB | |||
|
Domain Architecture |

|
|||||
| Description | 14-3-3 protein beta/alpha (Protein kinase C inhibitor protein 1) (KCIP-1) (Protein 1054). | |||||
|
NRP1_HUMAN
|
||||||
| NC score | 0.013805 (rank : 20) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.013504 (rank : 21) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
ZC11A_MOUSE
|
||||||
| NC score | 0.011828 (rank : 22) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.011660 (rank : 23) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
NOC4L_MOUSE
|
||||||
| NC score | 0.011532 (rank : 24) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHY2, Q8R1F7 | Gene names | Noc4l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 4 homolog (NOC4 protein homolog) (NOC4-like protein) (Nucleolar complex-associated protein 4-like protein). | |||||
|
TAF1L_HUMAN
|
||||||
| NC score | 0.011422 (rank : 25) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZX4 | Gene names | TAF1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID 210 kDa subunit (TBP-associated factor 210 kDa) (TAF(II)210) (TBP-associated factor 1-like). | |||||
|
CRNL1_MOUSE
|
||||||
| NC score | 0.011384 (rank : 26) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P63154, Q542E8, Q9CQC1 | Gene names | Crnkl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crooked neck-like protein 1 (Crooked neck homolog). | |||||
|
TAF1_HUMAN
|
||||||
| NC score | 0.011267 (rank : 27) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
UBP7_HUMAN
|
||||||
| NC score | 0.009634 (rank : 28) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
TRAP1_HUMAN
|
||||||
| NC score | 0.007744 (rank : 29) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12931, O43642, O75235, Q9UHL5 | Gene names | TRAP1, HSP75 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
BSSP4_MOUSE
|
||||||
| NC score | 0.007706 (rank : 30) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ER10 | Gene names | Prss22, Bssp4, Prss26 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific serine protease 4 precursor (EC 3.4.21.-) (BSSP-4) (Serine protease 22) (Tryptase epsilon). | |||||
|
DOC2A_HUMAN
|
||||||
| NC score | 0.007498 (rank : 31) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.007181 (rank : 32) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
ZFYV9_HUMAN
|
||||||
| NC score | 0.007175 (rank : 33) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
ABTB1_HUMAN
|
||||||
| NC score | 0.006962 (rank : 34) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969K4, Q6ZNU9, Q71MF1, Q96S62, Q96S63 | Gene names | ABTB1, BPOZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 1 (Elongation factor 1A-binding protein). | |||||
|
DOC10_HUMAN
|
||||||
| NC score | 0.006204 (rank : 35) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96BY6, O75178, Q9NW06, Q9NXI8 | Gene names | DOCK10, KIAA0694 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 10 (Protein zizimin 3). | |||||
|
BSSP4_HUMAN
|
||||||
| NC score | 0.004932 (rank : 36) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZN4, O43342, Q6UXE0 | Gene names | PRSS22, BSSP4, PRSS26 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific serine protease 4 precursor (EC 3.4.21.-) (BSSP-4) (Serine protease 22) (Tryptase epsilon). | |||||
|
KCC1D_MOUSE
|
||||||
| NC score | 0.004055 (rank : 37) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BW96, Q3U450, Q80W64, Q8BWI7 | Gene names | Camk1d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase type 1D (EC 2.7.11.17) (CaM kinase ID) (CaM kinase I delta) (CaMKI-delta) (CaM-KI delta) (CaMKI delta) (Camk1D) (CamKI-like protein kinase) (CKLiK) (mCKLiK). | |||||
|
KCC1D_HUMAN
|
||||||
| NC score | 0.004054 (rank : 38) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IU85, Q9HD31 | Gene names | CAMK1D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase type 1D (EC 2.7.11.17) (CaM kinase ID) (CaM kinase I delta) (CaMKI-delta) (CaM-KI delta) (CaMKI delta) (Camk1D) (CamKI-like protein kinase) (CKLiK). | |||||
|
K1HB_HUMAN
|
||||||
| NC score | 0.001755 (rank : 39) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14525, O76010 | Gene names | KRTHA3B, HHA3-II, HKA3B | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha3-II (Hair keratin, type I Ha3-II). | |||||