Please be patient as the page loads
|
CLH_MOUSE
|
||||||
| SwissProt Accessions | Q68FD5 | Gene names | Cltc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin heavy chain. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CLH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999327 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q00610, Q6N0A0, Q86TF2 | Gene names | CLTC, CLH17, KIAA0034 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 1 (CLH-17). | |||||
|
CLH2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995003 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
CLH_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q68FD5 | Gene names | Cltc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin heavy chain. | |||||
|
WDR35_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 4) | NC score | 0.075827 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2L0, Q8NE11 | Gene names | WDR35, KIAA1336 | |||
|
Domain Architecture |
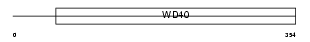
|
|||||
| Description | WD repeat protein 35. | |||||
|
NALD2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 5) | NC score | 0.027086 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3Q0 | Gene names | NAALAD2 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase 2 (EC 3.4.17.21) (N- acetylated-alpha-linked acidic dipeptidase II) (NAALADase II). | |||||
|
VPS41_HUMAN
|
||||||
| θ value | 0.813845 (rank : 6) | NC score | 0.083045 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49754, Q99851, Q99852 | Gene names | VPS41 | |||
|
Domain Architecture |
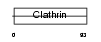
|
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (S53). | |||||
|
VPS41_MOUSE
|
||||||
| θ value | 0.813845 (rank : 7) | NC score | 0.083162 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5KU39, Q3TPS1, Q80V99, Q8BKK6, Q8CG01, Q8CJC3 | Gene names | Vps41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (VAM2 homolog) (mVAM2). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 8) | NC score | 0.012960 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
WDR35_MOUSE
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.068918 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BND3, Q8CDL4 | Gene names | Wdr35 | |||
|
Domain Architecture |
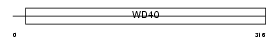
|
|||||
| Description | WD repeat protein 35. | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 10) | NC score | 0.008299 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
ZN613_HUMAN
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.001669 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PF04, Q96SS9 | Gene names | ZNF613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 613. | |||||
|
ADCY7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.012608 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51829 | Gene names | Adcy7 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
CTNA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.023123 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26232, Q4ZFW1, Q53R26, Q53R33, Q53T67, Q53T71, Q53TM8, Q7Z3Y0 | Gene names | CTNNA2, CAPR | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-2 (Alpha-catenin-related protein) (Alpha N-catenin). | |||||
|
CTNA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.023157 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61301, Q61300, Q6AXD1 | Gene names | Ctnna2, Catna2 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-2 (Alpha-catenin-related protein) (Alpha N-catenin). | |||||
|
KAD7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.036376 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96M32, Q8IYP6 | Gene names | AK7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
|
NRP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.018863 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.014683 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
UN45B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.020154 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWX7, Q495Q8, Q495Q9 | Gene names | UNC45B, CMYA4, UNC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
1433B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.016427 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31946 | Gene names | YWHAB | |||
|
Domain Architecture |
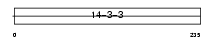
|
|||||
| Description | 14-3-3 protein beta/alpha (Protein kinase C inhibitor protein 1) (KCIP-1) (Protein 1054). | |||||
|
COPB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.009616 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O55029, Q3U5Z9 | Gene names | Copb2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta' (Beta'-coat protein) (Beta'-COP) (p102). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.008557 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
I2C2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.020106 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKV8, Q8TCZ5, Q8WV58, Q96ID1 | Gene names | EIF2C2, AGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 2 (eIF2C 2) (eIF-2C 2) (Argonaute-2) (Slicer protein) (PAZ Piwi domain protein) (PPD). | |||||
|
I2C2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.019966 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CJG0, Q4VAB3, Q571J6 | Gene names | Eif2c2, Ago2, Kiaa4215 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 2 (eIF2C 2) (eIF-2C 2) (Argonaute-2) (Slicer protein) (Piwi/argonaute family protein meIF2C2). | |||||
|
JAK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.003296 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52332, Q62126 | Gene names | Jak1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase JAK1 (EC 2.7.10.2) (Janus kinase 1) (JAK-1). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.006404 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
NRP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.015698 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.006545 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
ARHG5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.013519 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12774 | Gene names | ARHGEF5, TIM | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 5 (Guanine nucleotide regulatory protein TIM) (Oncogene TIM) (p60 TIM) (Transforming immortalized mammary oncogene). | |||||
|
BSSP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.005917 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ER10 | Gene names | Prss22, Bssp4, Prss26 | |||
|
Domain Architecture |
|
|||||
| Description | Brain-specific serine protease 4 precursor (EC 3.4.21.-) (BSSP-4) (Serine protease 22) (Tryptase epsilon). | |||||
|
KAD7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.028593 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D2H2, Q8BVH3 | Gene names | Ak7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.012033 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
DVL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.009856 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
JAK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.002257 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23458 | Gene names | JAK1, JAK1A | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase JAK1 (EC 2.7.10.2) (Janus kinase 1) (JAK-1). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.006224 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
SC24D_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.012912 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
UN45B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.015116 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
CLH_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q68FD5 | Gene names | Cltc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin heavy chain. | |||||
|
CLH1_HUMAN
|
||||||
| NC score | 0.999327 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q00610, Q6N0A0, Q86TF2 | Gene names | CLTC, CLH17, KIAA0034 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 1 (CLH-17). | |||||
|
CLH2_HUMAN
|
||||||
| NC score | 0.995003 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
VPS41_MOUSE
|
||||||
| NC score | 0.083162 (rank : 4) | θ value | 0.813845 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5KU39, Q3TPS1, Q80V99, Q8BKK6, Q8CG01, Q8CJC3 | Gene names | Vps41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (VAM2 homolog) (mVAM2). | |||||
|
VPS41_HUMAN
|
||||||
| NC score | 0.083045 (rank : 5) | θ value | 0.813845 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49754, Q99851, Q99852 | Gene names | VPS41 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (S53). | |||||
|
WDR35_HUMAN
|
||||||
| NC score | 0.075827 (rank : 6) | θ value | 0.0961366 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2L0, Q8NE11 | Gene names | WDR35, KIAA1336 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 35. | |||||
|
WDR35_MOUSE
|
||||||
| NC score | 0.068918 (rank : 7) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BND3, Q8CDL4 | Gene names | Wdr35 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 35. | |||||
|
KAD7_HUMAN
|
||||||
| NC score | 0.036376 (rank : 8) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96M32, Q8IYP6 | Gene names | AK7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
|
KAD7_MOUSE
|
||||||
| NC score | 0.028593 (rank : 9) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D2H2, Q8BVH3 | Gene names | Ak7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
|
NALD2_HUMAN
|
||||||
| NC score | 0.027086 (rank : 10) | θ value | 0.125558 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3Q0 | Gene names | NAALAD2 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase 2 (EC 3.4.17.21) (N- acetylated-alpha-linked acidic dipeptidase II) (NAALADase II). | |||||
|
CTNA2_MOUSE
|
||||||
| NC score | 0.023157 (rank : 11) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61301, Q61300, Q6AXD1 | Gene names | Ctnna2, Catna2 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-2 (Alpha-catenin-related protein) (Alpha N-catenin). | |||||
|
CTNA2_HUMAN
|
||||||
| NC score | 0.023123 (rank : 12) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26232, Q4ZFW1, Q53R26, Q53R33, Q53T67, Q53T71, Q53TM8, Q7Z3Y0 | Gene names | CTNNA2, CAPR | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-2 (Alpha-catenin-related protein) (Alpha N-catenin). | |||||
|
UN45B_HUMAN
|
||||||
| NC score | 0.020154 (rank : 13) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWX7, Q495Q8, Q495Q9 | Gene names | UNC45B, CMYA4, UNC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
I2C2_HUMAN
|
||||||
| NC score | 0.020106 (rank : 14) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKV8, Q8TCZ5, Q8WV58, Q96ID1 | Gene names | EIF2C2, AGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 2 (eIF2C 2) (eIF-2C 2) (Argonaute-2) (Slicer protein) (PAZ Piwi domain protein) (PPD). | |||||
|
I2C2_MOUSE
|
||||||
| NC score | 0.019966 (rank : 15) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CJG0, Q4VAB3, Q571J6 | Gene names | Eif2c2, Ago2, Kiaa4215 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 2 (eIF2C 2) (eIF-2C 2) (Argonaute-2) (Slicer protein) (Piwi/argonaute family protein meIF2C2). | |||||
|
NRP1_MOUSE
|
||||||
| NC score | 0.018863 (rank : 16) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
1433B_HUMAN
|
||||||
| NC score | 0.016427 (rank : 17) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31946 | Gene names | YWHAB | |||
|
Domain Architecture |

|
|||||
| Description | 14-3-3 protein beta/alpha (Protein kinase C inhibitor protein 1) (KCIP-1) (Protein 1054). | |||||
|
NRP1_HUMAN
|
||||||
| NC score | 0.015698 (rank : 18) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
UN45B_MOUSE
|
||||||
| NC score | 0.015116 (rank : 19) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.014683 (rank : 20) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
ARHG5_HUMAN
|
||||||
| NC score | 0.013519 (rank : 21) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12774 | Gene names | ARHGEF5, TIM | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 5 (Guanine nucleotide regulatory protein TIM) (Oncogene TIM) (p60 TIM) (Transforming immortalized mammary oncogene). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.012960 (rank : 22) | θ value | 1.38821 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
SC24D_HUMAN
|
||||||
| NC score | 0.012912 (rank : 23) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
ADCY7_MOUSE
|
||||||
| NC score | 0.012608 (rank : 24) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51829 | Gene names | Adcy7 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.012033 (rank : 25) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
DVL3_MOUSE
|
||||||
| NC score | 0.009856 (rank : 26) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
COPB2_MOUSE
|
||||||
| NC score | 0.009616 (rank : 27) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O55029, Q3U5Z9 | Gene names | Copb2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta' (Beta'-coat protein) (Beta'-COP) (p102). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.008557 (rank : 28) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.008299 (rank : 29) | θ value | 1.81305 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.006545 (rank : 30) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.006404 (rank : 31) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.006224 (rank : 32) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
BSSP4_MOUSE
|
||||||
| NC score | 0.005917 (rank : 33) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ER10 | Gene names | Prss22, Bssp4, Prss26 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific serine protease 4 precursor (EC 3.4.21.-) (BSSP-4) (Serine protease 22) (Tryptase epsilon). | |||||
|
JAK1_MOUSE
|
||||||
| NC score | 0.003296 (rank : 34) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52332, Q62126 | Gene names | Jak1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase JAK1 (EC 2.7.10.2) (Janus kinase 1) (JAK-1). | |||||
|
JAK1_HUMAN
|
||||||
| NC score | 0.002257 (rank : 35) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23458 | Gene names | JAK1, JAK1A | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase JAK1 (EC 2.7.10.2) (Janus kinase 1) (JAK-1). | |||||
|
ZN613_HUMAN
|
||||||
| NC score | 0.001669 (rank : 36) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PF04, Q96SS9 | Gene names | ZNF613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 613. | |||||