Please be patient as the page loads
|
VPS41_HUMAN
|
||||||
| SwissProt Accessions | P49754, Q99851, Q99852 | Gene names | VPS41 | |||
|
Domain Architecture |
|
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (S53). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
VPS41_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P49754, Q99851, Q99852 | Gene names | VPS41 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (S53). | |||||
|
VPS41_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996334 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5KU39, Q3TPS1, Q80V99, Q8BKK6, Q8CG01, Q8CJC3 | Gene names | Vps41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (VAM2 homolog) (mVAM2). | |||||
|
VPS11_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 3) | NC score | 0.245951 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H270, Q8WY89, Q96EP8, Q9H6D9, Q9HCS6 | Gene names | VPS11, RNF108 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 11 (hVPS11) (RING finger protein 108). | |||||
|
VPS18_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 4) | NC score | 0.246941 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R307, Q8BGV6, Q8BZX6, Q8C126 | Gene names | Vps18 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18. | |||||
|
VPS11_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 5) | NC score | 0.233967 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91W86, Q9DBX8 | Gene names | Vps11 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 11. | |||||
|
VPS18_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 6) | NC score | 0.241598 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P253, Q8TCG0, Q96DI3, Q9H268 | Gene names | VPS18, KIAA1475 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18 (hVPS18). | |||||
|
VPS39_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.099687 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R5L3, Q922I3 | Gene names | Vps39, Pldn | |||
|
Domain Architecture |

|
|||||
| Description | Vam6/Vps39-like protein (Vps39 protein). | |||||
|
WDR61_HUMAN
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.040381 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZS3, Q6IA22, Q7Z4X4 | Gene names | WDR61 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 61 (Meiotic recombination REC14 protein homolog). | |||||
|
WDR61_MOUSE
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.040444 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERF3, Q3U562, Q9CZP1 | Gene names | Wdr61 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 61 (Meiotic recombination REC14 protein homolog). | |||||
|
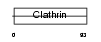
CLH2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.095128 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
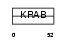
ZN267_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.005661 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14586, Q8NE41, Q9NRJ0 | Gene names | ZNF267 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 267 (Zinc finger protein HZF2). | |||||
|
I2C3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.041493 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H9G7, Q5TA55, Q9H1U6 | Gene names | EIF2C3, AGO3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 3 (eIF2C 3) (eIF-2C 3) (Argonaute-3). | |||||
|
I2C3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.041506 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CJF9 | Gene names | Eif2c3, Ago3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 3 (eIF2C 3) (eIF-2C 3) (Argonaute-3) (Piwi/argonaute family protein meIF2C3). | |||||
|
B3GL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.052790 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920V1, O54906, Q91V72, Q920V0, Q9CTE5 | Gene names | b3galnt1, B3galt3, B3gt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
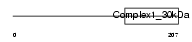
NDUS3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.095605 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75489, Q9UNQ8 | Gene names | NDUFS3 | |||
|
Domain Architecture |
|
|||||
| Description | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 30 kDa subunit) (Complex I-30kD) (CI-30kD). | |||||
|
CLH1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.083059 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00610, Q6N0A0, Q86TF2 | Gene names | CLTC, CLH17, KIAA0034 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 1 (CLH-17). | |||||
|
CLH_MOUSE
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.083045 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68FD5 | Gene names | Cltc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin heavy chain. | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.040381 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.018294 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
APEX1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.066375 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27695, Q969L5, Q99775 | Gene names | APEX1, APE, APEX, APX, HAP1, REF1 | |||
|
Domain Architecture |
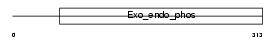
|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN) (REF-1 protein). | |||||
|
KAIN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.014192 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29622, Q53XB5, Q86TR9, Q96BZ5 | Gene names | SERPINA4, KST, PI4 | |||
|
Domain Architecture |
|
|||||
| Description | Kallistatin precursor (Serpin A4) (Kallikrein inhibitor) (Protease inhibitor 4). | |||||
|
APEX1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.062788 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28352 | Gene names | Apex1, Ape, Apex | |||
|
Domain Architecture |

|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN). | |||||
|
NDUS3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.075460 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCT2, Q8R073 | Gene names | Ndufs3 | |||
|
Domain Architecture |
|
|||||
| Description | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 30 kDa subunit) (Complex I-30kD) (CI-30kD). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.012397 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
B3GL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.039781 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75752, Q3Y531, Q6IAI5, Q8NFM8, Q9HA06 | Gene names | B3GALNT1, B3GALT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
TF3C3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.041886 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
I2C1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.034679 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UL18, Q5TA57, Q6P4S0 | Gene names | EIF2C1, AGO1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 1 (eIF2C 1) (eIF-2C 1) (Argonaute-1) (Putative RNA-binding protein Q99). | |||||
|
I2C1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.034736 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CJG1 | Gene names | Eif2c1, Ago1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 1 (eIF2C 1) (eIF-2C 1) (Argonaute-1) (Piwi/argonaute family protein meIF2C1). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.025271 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
CD83_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.045852 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01151 | Gene names | CD83 | |||
|
Domain Architecture |
|
|||||
| Description | CD83 antigen precursor (Cell surface protein HB15) (B-cell activation protein). | |||||
|
HS90A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.016617 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07900, Q2PP14, Q9BVQ5 | Gene names | HSP90AA1, HSP90A, HSPC1, HSPCA | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (NY-REN-38 antigen). | |||||
|
SART2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.024284 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BLI4, Q3U620 | Gene names | Sart2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 2 precursor (SART-2). | |||||
|
TRUB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.017283 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C0D0, Q5EBH7, Q9D331 | Gene names | Trub1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tRNA pseudouridine synthase 1 (EC 5.4.99.-). | |||||
|
UB7I3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.013554 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUB0 | Gene names | Rbck1, Rbck, Ubce7ip3, Uip28 | |||
|
Domain Architecture |
|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (UbcM4-interacting protein 28). | |||||
|
RN126_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.011352 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YL2 | Gene names | Rnf126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
STIM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.011651 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
TRPA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.011500 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
VPS41_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P49754, Q99851, Q99852 | Gene names | VPS41 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (S53). | |||||
|
VPS41_MOUSE
|
||||||
| NC score | 0.996334 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5KU39, Q3TPS1, Q80V99, Q8BKK6, Q8CG01, Q8CJC3 | Gene names | Vps41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (VAM2 homolog) (mVAM2). | |||||
|
VPS18_MOUSE
|
||||||
| NC score | 0.246941 (rank : 3) | θ value | 9.29e-05 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R307, Q8BGV6, Q8BZX6, Q8C126 | Gene names | Vps18 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18. | |||||
|
VPS11_HUMAN
|
||||||
| NC score | 0.245951 (rank : 4) | θ value | 3.19293e-05 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H270, Q8WY89, Q96EP8, Q9H6D9, Q9HCS6 | Gene names | VPS11, RNF108 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 11 (hVPS11) (RING finger protein 108). | |||||
|
VPS18_HUMAN
|
||||||
| NC score | 0.241598 (rank : 5) | θ value | 0.00020696 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P253, Q8TCG0, Q96DI3, Q9H268 | Gene names | VPS18, KIAA1475 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18 (hVPS18). | |||||
|
VPS11_MOUSE
|
||||||
| NC score | 0.233967 (rank : 6) | θ value | 0.000158464 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91W86, Q9DBX8 | Gene names | Vps11 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 11. | |||||
|
VPS39_MOUSE
|
||||||
| NC score | 0.099687 (rank : 7) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R5L3, Q922I3 | Gene names | Vps39, Pldn | |||
|
Domain Architecture |

|
|||||
| Description | Vam6/Vps39-like protein (Vps39 protein). | |||||
|
NDUS3_HUMAN
|
||||||
| NC score | 0.095605 (rank : 8) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75489, Q9UNQ8 | Gene names | NDUFS3 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 30 kDa subunit) (Complex I-30kD) (CI-30kD). | |||||
|
CLH2_HUMAN
|
||||||
| NC score | 0.095128 (rank : 9) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
CLH1_HUMAN
|
||||||
| NC score | 0.083059 (rank : 10) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00610, Q6N0A0, Q86TF2 | Gene names | CLTC, CLH17, KIAA0034 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 1 (CLH-17). | |||||
|
CLH_MOUSE
|
||||||
| NC score | 0.083045 (rank : 11) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68FD5 | Gene names | Cltc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin heavy chain. | |||||
|
NDUS3_MOUSE
|
||||||
| NC score | 0.075460 (rank : 12) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCT2, Q8R073 | Gene names | Ndufs3 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 30 kDa subunit) (Complex I-30kD) (CI-30kD). | |||||
|
APEX1_HUMAN
|
||||||
| NC score | 0.066375 (rank : 13) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27695, Q969L5, Q99775 | Gene names | APEX1, APE, APEX, APX, HAP1, REF1 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN) (REF-1 protein). | |||||
|
APEX1_MOUSE
|
||||||
| NC score | 0.062788 (rank : 14) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28352 | Gene names | Apex1, Ape, Apex | |||
|
Domain Architecture |

|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN). | |||||
|
B3GL1_MOUSE
|
||||||
| NC score | 0.052790 (rank : 15) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920V1, O54906, Q91V72, Q920V0, Q9CTE5 | Gene names | b3galnt1, B3galt3, B3gt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
CD83_HUMAN
|
||||||
| NC score | 0.045852 (rank : 16) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01151 | Gene names | CD83 | |||
|
Domain Architecture |

|
|||||
| Description | CD83 antigen precursor (Cell surface protein HB15) (B-cell activation protein). | |||||
|
TF3C3_HUMAN
|
||||||
| NC score | 0.041886 (rank : 17) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
I2C3_MOUSE
|
||||||
| NC score | 0.041506 (rank : 18) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CJF9 | Gene names | Eif2c3, Ago3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 3 (eIF2C 3) (eIF-2C 3) (Argonaute-3) (Piwi/argonaute family protein meIF2C3). | |||||
|
I2C3_HUMAN
|
||||||
| NC score | 0.041493 (rank : 19) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H9G7, Q5TA55, Q9H1U6 | Gene names | EIF2C3, AGO3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 3 (eIF2C 3) (eIF-2C 3) (Argonaute-3). | |||||
|
WDR61_MOUSE
|
||||||
| NC score | 0.040444 (rank : 20) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERF3, Q3U562, Q9CZP1 | Gene names | Wdr61 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 61 (Meiotic recombination REC14 protein homolog). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.040381 (rank : 21) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
WDR61_HUMAN
|
||||||
| NC score | 0.040381 (rank : 22) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZS3, Q6IA22, Q7Z4X4 | Gene names | WDR61 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 61 (Meiotic recombination REC14 protein homolog). | |||||
|
B3GL1_HUMAN
|
||||||
| NC score | 0.039781 (rank : 23) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75752, Q3Y531, Q6IAI5, Q8NFM8, Q9HA06 | Gene names | B3GALNT1, B3GALT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
I2C1_MOUSE
|
||||||
| NC score | 0.034736 (rank : 24) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CJG1 | Gene names | Eif2c1, Ago1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 1 (eIF2C 1) (eIF-2C 1) (Argonaute-1) (Piwi/argonaute family protein meIF2C1). | |||||
|
I2C1_HUMAN
|
||||||
| NC score | 0.034679 (rank : 25) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UL18, Q5TA57, Q6P4S0 | Gene names | EIF2C1, AGO1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2C 1 (eIF2C 1) (eIF-2C 1) (Argonaute-1) (Putative RNA-binding protein Q99). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.025271 (rank : 26) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
SART2_MOUSE
|
||||||
| NC score | 0.024284 (rank : 27) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BLI4, Q3U620 | Gene names | Sart2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 2 precursor (SART-2). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.018294 (rank : 28) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
TRUB1_MOUSE
|
||||||
| NC score | 0.017283 (rank : 29) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C0D0, Q5EBH7, Q9D331 | Gene names | Trub1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tRNA pseudouridine synthase 1 (EC 5.4.99.-). | |||||
|
HS90A_HUMAN
|
||||||
| NC score | 0.016617 (rank : 30) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07900, Q2PP14, Q9BVQ5 | Gene names | HSP90AA1, HSP90A, HSPC1, HSPCA | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (NY-REN-38 antigen). | |||||
|
KAIN_HUMAN
|
||||||
| NC score | 0.014192 (rank : 31) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29622, Q53XB5, Q86TR9, Q96BZ5 | Gene names | SERPINA4, KST, PI4 | |||
|
Domain Architecture |

|
|||||
| Description | Kallistatin precursor (Serpin A4) (Kallikrein inhibitor) (Protease inhibitor 4). | |||||
|
UB7I3_MOUSE
|
||||||
| NC score | 0.013554 (rank : 32) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUB0 | Gene names | Rbck1, Rbck, Ubce7ip3, Uip28 | |||
|
Domain Architecture |

|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (UbcM4-interacting protein 28). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.012397 (rank : 33) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
STIM1_HUMAN
|
||||||
| NC score | 0.011651 (rank : 34) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
TRPA1_MOUSE
|
||||||
| NC score | 0.011500 (rank : 35) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
RN126_MOUSE
|
||||||
| NC score | 0.011352 (rank : 36) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YL2 | Gene names | Rnf126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
ZN267_HUMAN
|
||||||
| NC score | 0.005661 (rank : 37) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14586, Q8NE41, Q9NRJ0 | Gene names | ZNF267 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 267 (Zinc finger protein HZF2). | |||||