Please be patient as the page loads
|
APEX1_HUMAN
|
||||||
| SwissProt Accessions | P27695, Q969L5, Q99775 | Gene names | APEX1, APE, APEX, APX, HAP1, REF1 | |||
|
Domain Architecture |
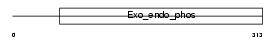
|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN) (REF-1 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
APEX1_HUMAN
|
||||||
| θ value | 4.38123e-180 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27695, Q969L5, Q99775 | Gene names | APEX1, APE, APEX, APX, HAP1, REF1 | |||
|
Domain Architecture |
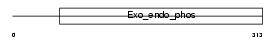
|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN) (REF-1 protein). | |||||
|
APEX1_MOUSE
|
||||||
| θ value | 1.08164e-170 (rank : 2) | NC score | 0.997663 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28352 | Gene names | Apex1, Ape, Apex | |||
|
Domain Architecture |

|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN). | |||||
|
APEX2_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 3) | NC score | 0.606003 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q68G58, Q8BJP7, Q8BTR7, Q8BUZ2, Q8BYE9, Q8R018, Q8R328, Q9CS12 | Gene names | Apex2, Ape2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2). | |||||
|
APEX2_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 4) | NC score | 0.590288 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBZ4, Q9Y5X7 | Gene names | APEX2, APE2, APEXL2, XTH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2) (APEX nuclease-like 2) (AP endonuclease XTH2). | |||||
|
LIN1_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 5) | NC score | 0.288087 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08547 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LINE-1 reverse transcriptase homolog. | |||||
|
POL2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 6) | NC score | 0.210660 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11369, Q60713, Q61787 | Gene names | Pol | |||
|
Domain Architecture |

|
|||||
| Description | Retrovirus-related Pol polyprotein LINE-1 (Long interspersed element- 1) (L1) [Includes: Reverse transcriptase (EC 2.7.7.49); Endonuclease]. | |||||
|
VPS41_HUMAN
|
||||||
| θ value | 1.81305 (rank : 7) | NC score | 0.066375 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49754, Q99851, Q99852 | Gene names | VPS41 | |||
|
Domain Architecture |
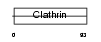
|
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (S53). | |||||
|
VPS41_MOUSE
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.066466 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5KU39, Q3TPS1, Q80V99, Q8BKK6, Q8CG01, Q8CJC3 | Gene names | Vps41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (VAM2 homolog) (mVAM2). | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 9) | NC score | 0.030899 (rank : 10) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
NEIL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 10) | NC score | 0.056055 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
APEX1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.38123e-180 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27695, Q969L5, Q99775 | Gene names | APEX1, APE, APEX, APX, HAP1, REF1 | |||
|
Domain Architecture |
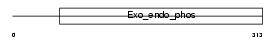
|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN) (REF-1 protein). | |||||
|
APEX1_MOUSE
|
||||||
| NC score | 0.997663 (rank : 2) | θ value | 1.08164e-170 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28352 | Gene names | Apex1, Ape, Apex | |||
|
Domain Architecture |

|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN). | |||||
|
APEX2_MOUSE
|
||||||
| NC score | 0.606003 (rank : 3) | θ value | 2.7842e-17 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q68G58, Q8BJP7, Q8BTR7, Q8BUZ2, Q8BYE9, Q8R018, Q8R328, Q9CS12 | Gene names | Apex2, Ape2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2). | |||||
|
APEX2_HUMAN
|
||||||
| NC score | 0.590288 (rank : 4) | θ value | 6.85773e-16 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBZ4, Q9Y5X7 | Gene names | APEX2, APE2, APEXL2, XTH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2) (APEX nuclease-like 2) (AP endonuclease XTH2). | |||||
|
LIN1_HUMAN
|
||||||
| NC score | 0.288087 (rank : 5) | θ value | 4.92598e-06 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08547 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LINE-1 reverse transcriptase homolog. | |||||
|
POL2_MOUSE
|
||||||
| NC score | 0.210660 (rank : 6) | θ value | 0.0330416 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11369, Q60713, Q61787 | Gene names | Pol | |||
|
Domain Architecture |

|
|||||
| Description | Retrovirus-related Pol polyprotein LINE-1 (Long interspersed element- 1) (L1) [Includes: Reverse transcriptase (EC 2.7.7.49); Endonuclease]. | |||||
|
VPS41_MOUSE
|
||||||
| NC score | 0.066466 (rank : 7) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5KU39, Q3TPS1, Q80V99, Q8BKK6, Q8CG01, Q8CJC3 | Gene names | Vps41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (VAM2 homolog) (mVAM2). | |||||
|
VPS41_HUMAN
|
||||||
| NC score | 0.066375 (rank : 8) | θ value | 1.81305 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49754, Q99851, Q99852 | Gene names | VPS41 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 41 homolog (S53). | |||||
|
NEIL3_MOUSE
|
||||||
| NC score | 0.056055 (rank : 9) | θ value | θ > 10 (rank : 10) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.030899 (rank : 10) | θ value | 6.88961 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||