Please be patient as the page loads
|
APEX2_HUMAN
|
||||||
| SwissProt Accessions | Q9UBZ4, Q9Y5X7 | Gene names | APEX2, APE2, APEXL2, XTH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2) (APEX nuclease-like 2) (AP endonuclease XTH2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
APEX2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UBZ4, Q9Y5X7 | Gene names | APEX2, APE2, APEXL2, XTH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2) (APEX nuclease-like 2) (AP endonuclease XTH2). | |||||
|
APEX2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994768 (rank : 2) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q68G58, Q8BJP7, Q8BTR7, Q8BUZ2, Q8BYE9, Q8R018, Q8R328, Q9CS12 | Gene names | Apex2, Ape2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2). | |||||
|
APEX1_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 3) | NC score | 0.590288 (rank : 3) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27695, Q969L5, Q99775 | Gene names | APEX1, APE, APEX, APX, HAP1, REF1 | |||
|
Domain Architecture |
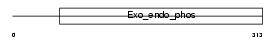
|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN) (REF-1 protein). | |||||
|
APEX1_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 4) | NC score | 0.587422 (rank : 4) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P28352 | Gene names | Apex1, Ape, Apex | |||
|
Domain Architecture |

|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN). | |||||
|
NEIL3_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 5) | NC score | 0.329214 (rank : 5) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
LIN1_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 6) | NC score | 0.218120 (rank : 9) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08547 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LINE-1 reverse transcriptase homolog. | |||||
|
NEIL3_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 7) | NC score | 0.307134 (rank : 6) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
TOP3A_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 8) | NC score | 0.239307 (rank : 7) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70157 | Gene names | Top3a, Top3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
TOP3A_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 9) | NC score | 0.230853 (rank : 8) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13472, Q13473 | Gene names | TOP3A, TOP3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.027864 (rank : 15) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 11) | NC score | 0.018076 (rank : 17) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.025334 (rank : 16) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.047759 (rank : 13) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.013241 (rank : 19) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
BC11B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 15) | NC score | 0.001685 (rank : 21) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 767 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PV8, Q8C2I1, Q99PV6, Q99PV7, Q9JLF8 | Gene names | Bcl11b, Ctip2, Rit1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma/leukemia 11B (B-cell CLL/lymphoma 11B) (Radiation- induced tumor suppressor gene 1 protein) (mRit1) (COUP-TF-interacting protein 2). | |||||
|
IDH3A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.014433 (rank : 18) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50213, Q9H3X0 | Gene names | IDH3A | |||
|
Domain Architecture |
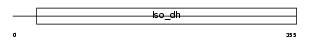
|
|||||
| Description | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial precursor (EC 1.1.1.41) (Isocitric dehydrogenase) (NAD(+)-specific ICDH). | |||||
|
TTF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.035409 (rank : 14) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.008538 (rank : 20) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
POL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 19) | NC score | 0.116962 (rank : 10) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11369, Q60713, Q61787 | Gene names | Pol | |||
|
Domain Architecture |

|
|||||
| Description | Retrovirus-related Pol polyprotein LINE-1 (Long interspersed element- 1) (L1) [Includes: Reverse transcriptase (EC 2.7.7.49); Endonuclease]. | |||||
|
TOP3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 20) | NC score | 0.074157 (rank : 12) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95985, Q9BUP5 | Gene names | TOP3B, TOP3B1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-beta-1 (EC 5.99.1.2) (DNA topoisomerase III beta- 1). | |||||
|
TOP3B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 21) | NC score | 0.074399 (rank : 11) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z321 | Gene names | Top3b, Top3b1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-beta-1 (EC 5.99.1.2) (DNA topoisomerase III beta- 1). | |||||
|
APEX2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UBZ4, Q9Y5X7 | Gene names | APEX2, APE2, APEXL2, XTH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2) (APEX nuclease-like 2) (AP endonuclease XTH2). | |||||
|
APEX2_MOUSE
|
||||||
| NC score | 0.994768 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q68G58, Q8BJP7, Q8BTR7, Q8BUZ2, Q8BYE9, Q8R018, Q8R328, Q9CS12 | Gene names | Apex2, Ape2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2). | |||||
|
APEX1_HUMAN
|
||||||
| NC score | 0.590288 (rank : 3) | θ value | 6.85773e-16 (rank : 3) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27695, Q969L5, Q99775 | Gene names | APEX1, APE, APEX, APX, HAP1, REF1 | |||
|
Domain Architecture |
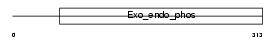
|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN) (REF-1 protein). | |||||
|
APEX1_MOUSE
|
||||||
| NC score | 0.587422 (rank : 4) | θ value | 1.99529e-15 (rank : 4) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P28352 | Gene names | Apex1, Ape, Apex | |||
|
Domain Architecture |

|
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase (EC 4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN). | |||||
|
NEIL3_MOUSE
|
||||||
| NC score | 0.329214 (rank : 5) | θ value | 7.59969e-07 (rank : 5) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
NEIL3_HUMAN
|
||||||
| NC score | 0.307134 (rank : 6) | θ value | 2.44474e-05 (rank : 7) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
TOP3A_MOUSE
|
||||||
| NC score | 0.239307 (rank : 7) | θ value | 2.44474e-05 (rank : 8) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70157 | Gene names | Top3a, Top3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
TOP3A_HUMAN
|
||||||
| NC score | 0.230853 (rank : 8) | θ value | 0.000121331 (rank : 9) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13472, Q13473 | Gene names | TOP3A, TOP3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
LIN1_HUMAN
|
||||||
| NC score | 0.218120 (rank : 9) | θ value | 1.09739e-05 (rank : 6) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08547 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LINE-1 reverse transcriptase homolog. | |||||
|
POL2_MOUSE
|
||||||
| NC score | 0.116962 (rank : 10) | θ value | θ > 10 (rank : 19) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11369, Q60713, Q61787 | Gene names | Pol | |||
|
Domain Architecture |

|
|||||
| Description | Retrovirus-related Pol polyprotein LINE-1 (Long interspersed element- 1) (L1) [Includes: Reverse transcriptase (EC 2.7.7.49); Endonuclease]. | |||||
|
TOP3B_MOUSE
|
||||||
| NC score | 0.074399 (rank : 11) | θ value | θ > 10 (rank : 21) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z321 | Gene names | Top3b, Top3b1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-beta-1 (EC 5.99.1.2) (DNA topoisomerase III beta- 1). | |||||
|
TOP3B_HUMAN
|
||||||
| NC score | 0.074157 (rank : 12) | θ value | θ > 10 (rank : 20) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95985, Q9BUP5 | Gene names | TOP3B, TOP3B1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-beta-1 (EC 5.99.1.2) (DNA topoisomerase III beta- 1). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.047759 (rank : 13) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
TTF2_HUMAN
|
||||||
| NC score | 0.035409 (rank : 14) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.027864 (rank : 15) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.025334 (rank : 16) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.018076 (rank : 17) | θ value | 0.62314 (rank : 11) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
IDH3A_HUMAN
|
||||||
| NC score | 0.014433 (rank : 18) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50213, Q9H3X0 | Gene names | IDH3A | |||
|
Domain Architecture |

|
|||||
| Description | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial precursor (EC 1.1.1.41) (Isocitric dehydrogenase) (NAD(+)-specific ICDH). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.013241 (rank : 19) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.008538 (rank : 20) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
BC11B_MOUSE
|
||||||
| NC score | 0.001685 (rank : 21) | θ value | 6.88961 (rank : 15) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 767 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PV8, Q8C2I1, Q99PV6, Q99PV7, Q9JLF8 | Gene names | Bcl11b, Ctip2, Rit1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma/leukemia 11B (B-cell CLL/lymphoma 11B) (Radiation- induced tumor suppressor gene 1 protein) (mRit1) (COUP-TF-interacting protein 2). | |||||