Please be patient as the page loads
|
TTF2_HUMAN
|
||||||
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TTF2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.940595 (rank : 2) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
SMRA3_HUMAN
|
||||||
| θ value | 2.24614e-67 (rank : 3) | NC score | 0.809813 (rank : 25) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14527, Q14536, Q16051, Q7KYJ6, Q86YA5, Q92652, Q96KM9 | Gene names | SMARCA3, HIP116A, HLTF, SNF2L3, ZBU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (DNA-binding protein/plasminogen activator inhibitor 1 regulator) (Helicase-like transcription factor) (HIP116). | |||||
|
SMRA3_MOUSE
|
||||||
| θ value | 4.84666e-62 (rank : 4) | NC score | 0.813398 (rank : 24) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6PCN7, O35596, O35597, Q80VT6 | Gene names | Smarca3, Snf2l3, Zbu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (TNF-response element-binding protein) (P113). | |||||
|
RA54B_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 5) | NC score | 0.874486 (rank : 4) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y620 | Gene names | RAD54B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
RA54B_MOUSE
|
||||||
| θ value | 3.61106e-41 (rank : 6) | NC score | 0.871769 (rank : 7) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6PFE3 | Gene names | Rad54b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
SMCA1_MOUSE
|
||||||
| θ value | 2.34062e-40 (rank : 7) | NC score | 0.859726 (rank : 11) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
SMCA1_HUMAN
|
||||||
| θ value | 2.58786e-39 (rank : 8) | NC score | 0.861380 (rank : 8) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P28370, Q5JV41, Q5JV42 | Gene names | SMARCA1, SNF2L, SNF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1). | |||||
|
SMCA5_MOUSE
|
||||||
| θ value | 4.13158e-37 (rank : 9) | NC score | 0.860705 (rank : 9) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91ZW3, Q8C791, Q8CA22, Q8VDG1, Q925M9 | Gene names | Smarca5, Snf2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (Sucrose nonfermenting protein 2 homolog) (mSnf2h). | |||||
|
SMCA5_HUMAN
|
||||||
| θ value | 9.20422e-37 (rank : 10) | NC score | 0.860368 (rank : 10) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60264 | Gene names | SMARCA5, SNF2H, WCRF135 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin A5) (Sucrose nonfermenting protein 2 homolog) (hSNF2H). | |||||
|
ERCC6_HUMAN
|
||||||
| θ value | 1.32909e-35 (rank : 11) | NC score | 0.859580 (rank : 12) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
SMRCD_HUMAN
|
||||||
| θ value | 3.86705e-35 (rank : 12) | NC score | 0.874938 (rank : 3) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
|
BTAF1_HUMAN
|
||||||
| θ value | 1.12514e-34 (rank : 13) | NC score | 0.873290 (rank : 5) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 14) | NC score | 0.831350 (rank : 15) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
SMRCD_MOUSE
|
||||||
| θ value | 3.3877e-31 (rank : 15) | NC score | 0.872378 (rank : 6) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q04692 | Gene names | Smarcad1, Etl1, Kiaa1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase SMARCAD1) (Enhancer trap locus homolog 1) (Etl-1). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 2.86786e-30 (rank : 16) | NC score | 0.829498 (rank : 16) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 3.74554e-30 (rank : 17) | NC score | 0.824725 (rank : 20) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 18) | NC score | 0.814292 (rank : 23) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 6.38894e-30 (rank : 19) | NC score | 0.818897 (rank : 22) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 2.42779e-29 (rank : 20) | NC score | 0.824640 (rank : 21) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 2.42779e-29 (rank : 21) | NC score | 0.827532 (rank : 19) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 4.14116e-29 (rank : 22) | NC score | 0.805813 (rank : 26) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 6.61148e-27 (rank : 23) | NC score | 0.792921 (rank : 27) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
RAD54_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 24) | NC score | 0.858056 (rank : 13) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92698, Q6IUY3 | Gene names | RAD54L, RAD54A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (hRAD54) (hHR54). | |||||
|
INOC1_HUMAN
|
||||||
| θ value | 1.5242e-23 (rank : 25) | NC score | 0.828505 (rank : 17) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
INOC1_MOUSE
|
||||||
| θ value | 3.39556e-23 (rank : 26) | NC score | 0.827995 (rank : 18) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6ZPV2, Q6P7V0, Q6PCP1, Q8C9T7 | Gene names | Inoc1, Kiaa1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-). | |||||
|
RAD54_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 27) | NC score | 0.854351 (rank : 14) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70270, Q8BSR5, Q8C2C4, Q8K3D4 | Gene names | Rad54l, Rad54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (mRAD54) (mHR54). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 28) | NC score | 0.746341 (rank : 30) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 29) | NC score | 0.743306 (rank : 33) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 30) | NC score | 0.744549 (rank : 32) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 31) | NC score | 0.745229 (rank : 31) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 32) | NC score | 0.661297 (rank : 36) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 1.16975e-15 (rank : 33) | NC score | 0.722482 (rank : 35) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 34) | NC score | 0.641503 (rank : 37) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 35) | NC score | 0.728147 (rank : 34) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 36) | NC score | 0.765400 (rank : 29) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
SMAL1_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 37) | NC score | 0.771264 (rank : 28) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
NEIL3_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 38) | NC score | 0.105243 (rank : 38) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 39) | NC score | 0.038094 (rank : 72) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NEIL3_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 40) | NC score | 0.098608 (rank : 39) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 41) | NC score | 0.020120 (rank : 89) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
ZN451_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 42) | NC score | 0.019859 (rank : 90) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 627 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4E5, Q5VVF1, Q8N380, Q8TD15, Q9NQM1 | Gene names | ZNF451, COASTER, KIAA0576 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 451 (Coactivator for steroid receptors). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 43) | NC score | 0.033387 (rank : 78) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
NPDC1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 44) | NC score | 0.044240 (rank : 71) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64322 | Gene names | Npdc1, Npdc-1 | |||
|
Domain Architecture |
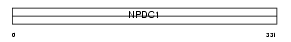
|
|||||
| Description | Neural proliferation differentiation and control protein 1 precursor (NPDC-1 protein). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 45) | NC score | 0.028496 (rank : 81) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
CNGB3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 46) | NC score | 0.017910 (rank : 94) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
SNRK_MOUSE
|
||||||
| θ value | 0.21417 (rank : 47) | NC score | 0.000017 (rank : 124) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDU5, Q91WX6 | Gene names | Snrk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SNF-related serine/threonine-protein kinase (EC 2.7.11.1) (SNF1- related kinase). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.014908 (rank : 100) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
NFAT5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.018519 (rank : 93) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
SNRK_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | -0.000377 (rank : 125) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRH2, Q14706, Q68D15, Q6IQ46, Q9NXI7 | Gene names | SNRK, KIAA0096, SNFRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SNF-related serine/threonine-protein kinase (EC 2.7.11.1) (SNF1- related kinase). | |||||
|
TRI29_HUMAN
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.020261 (rank : 88) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14134, Q96AA9, Q9BZY7 | Gene names | TRIM29, ATDC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 29 (Ataxia-telangiectasia group D- associated protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.067162 (rank : 41) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.019487 (rank : 91) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
ZN451_MOUSE
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.015454 (rank : 99) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0P7 | Gene names | Znf451, Zfp451 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 451. | |||||
|
WDHD1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.012439 (rank : 106) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59328, Q6P408 | Gene names | Wdhd1, And1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
DHX34_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.008578 (rank : 115) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
EZRI_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.009924 (rank : 114) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.030030 (rank : 80) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
TOP3A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.037408 (rank : 74) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13472, Q13473 | Gene names | TOP3A, TOP3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.023045 (rank : 85) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
MGAT5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.026461 (rank : 83) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q09328 | Gene names | MGAT5, GGNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V (EC 2.4.1.155) (Mannoside acetylglucosaminyltransferase 5) (Alpha- mannoside beta-1,6-N-acetylglucosaminyltransferase) (N- acetylglucosaminyl-transferase V) (GNT-V) (GlcNAc-T V). | |||||
|
MGAT5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.026441 (rank : 84) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4G6 | Gene names | Mgat5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V (EC 2.4.1.155) (Mannoside acetylglucosaminyltransferase 5) (Alpha- mannoside beta-1,6-N-acetylglucosaminyltransferase) (N- acetylglucosaminyl-transferase V) (GNT-V) (GlcNAc-T V). | |||||
|
TCEA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.011946 (rank : 108) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23193, Q15563, Q6FG87 | Gene names | TCEA1, TFIIS | |||
|
Domain Architecture |
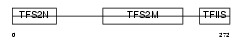
|
|||||
| Description | Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o). | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.012611 (rank : 104) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
DPOLA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.016168 (rank : 96) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33609 | Gene names | Pola1, Pola | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.036423 (rank : 75) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.022591 (rank : 86) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.015796 (rank : 97) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ZN690_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | -0.003547 (rank : 127) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IWY8, Q32M75, Q32M76, Q8NA40 | Gene names | ZNF690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 690. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.012337 (rank : 107) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.027580 (rank : 82) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ZBT40_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | -0.000720 (rank : 126) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
ATF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.011059 (rank : 112) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
EFHC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.019355 (rank : 92) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JST6, Q5JST8, Q68DK4, Q8NEI0, Q9H653 | Gene names | EFHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member C2. | |||||
|
GP179_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.020940 (rank : 87) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.013824 (rank : 101) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
SIA7A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.011537 (rank : 110) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZ39, Q9JJP5 | Gene names | St6galnac1, Siat7a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
STX12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.011832 (rank : 109) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86Y82, O95564 | Gene names | STX12 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-12. | |||||
|
UACA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.008093 (rank : 116) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
ZW10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.013012 (rank : 102) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43264 | Gene names | ZW10 | |||
|
Domain Architecture |

|
|||||
| Description | Centromere/kinetochore protein zw10 homolog. | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.006053 (rank : 120) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
O2T33_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | -0.004220 (rank : 128) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NG76 | Gene names | OR2T33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 2T33 (Olfactory receptor OR1-56). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.006453 (rank : 119) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.016356 (rank : 95) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
APEX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.035409 (rank : 76) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UBZ4, Q9Y5X7 | Gene names | APEX2, APE2, APEXL2, XTH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2) (APEX nuclease-like 2) (AP endonuclease XTH2). | |||||
|
APEX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.033234 (rank : 79) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68G58, Q8BJP7, Q8BTR7, Q8BUZ2, Q8BYE9, Q8R018, Q8R328, Q9CS12 | Gene names | Apex2, Ape2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2). | |||||
|
CF064_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.015482 (rank : 98) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K190 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf64 homolog. | |||||
|
DEN2A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.005566 (rank : 121) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C4S8, Q3TJU2, Q3TUG3, Q3UXA3 | Gene names | Dennd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
HASP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.012699 (rank : 103) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0R0, Q99MU9, Q9JJJ4 | Gene names | Gsg2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Haspin (EC 2.7.11.1) (Haploid germ cell-specific nuclear protein kinase) (Germ cell-specific gene 2 protein). | |||||
|
STX12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.011063 (rank : 111) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ER00, Q3UIV9, Q921T9 | Gene names | Stx12 | |||
|
Domain Architecture |
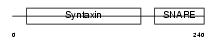
|
|||||
| Description | Syntaxin-12. | |||||
|
TOP3A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.034838 (rank : 77) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70157 | Gene names | Top3a, Top3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.037707 (rank : 73) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
DPOLA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.012480 (rank : 105) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09884, Q86UQ7 | Gene names | POLA1, POLA | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.006460 (rank : 118) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
POT14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.000992 (rank : 123) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6S5H5, Q6S5H6, Q6S8J2 | Gene names | POTE14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 14. | |||||
|
SPC25_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.011005 (rank : 113) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UA16, Q9D021, Q9D1K6 | Gene names | Spbc25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25. | |||||
|
TUFT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.007282 (rank : 117) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
WWP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.001297 (rank : 122) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.064091 (rank : 45) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.065254 (rank : 42) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.050532 (rank : 70) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.053769 (rank : 64) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.067338 (rank : 40) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CBX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.052074 (rank : 66) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.052074 (rank : 67) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.060603 (rank : 49) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.053284 (rank : 65) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.060704 (rank : 48) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.054027 (rank : 63) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.056579 (rank : 56) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.058788 (rank : 51) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.059083 (rank : 50) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.054349 (rank : 61) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.055267 (rank : 59) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.054103 (rank : 62) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.056257 (rank : 58) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.061535 (rank : 47) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.061902 (rank : 46) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.064821 (rank : 43) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.057361 (rank : 53) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.058261 (rank : 52) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.056662 (rank : 55) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.051178 (rank : 69) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
SP110_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.051319 (rank : 68) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.064414 (rank : 44) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.056912 (rank : 54) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.056487 (rank : 57) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.054504 (rank : 60) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
TTF2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.940595 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
SMRCD_HUMAN
|
||||||
| NC score | 0.874938 (rank : 3) | θ value | 3.86705e-35 (rank : 12) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
|
RA54B_HUMAN
|
||||||
| NC score | 0.874486 (rank : 4) | θ value | 3.85808e-43 (rank : 5) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y620 | Gene names | RAD54B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
BTAF1_HUMAN
|
||||||
| NC score | 0.873290 (rank : 5) | θ value | 1.12514e-34 (rank : 13) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
SMRCD_MOUSE
|
||||||
| NC score | 0.872378 (rank : 6) | θ value | 3.3877e-31 (rank : 15) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q04692 | Gene names | Smarcad1, Etl1, Kiaa1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase SMARCAD1) (Enhancer trap locus homolog 1) (Etl-1). | |||||
|
RA54B_MOUSE
|
||||||
| NC score | 0.871769 (rank : 7) | θ value | 3.61106e-41 (rank : 6) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6PFE3 | Gene names | Rad54b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
SMCA1_HUMAN
|
||||||
| NC score | 0.861380 (rank : 8) | θ value | 2.58786e-39 (rank : 8) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P28370, Q5JV41, Q5JV42 | Gene names | SMARCA1, SNF2L, SNF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1). | |||||
|
SMCA5_MOUSE
|
||||||
| NC score | 0.860705 (rank : 9) | θ value | 4.13158e-37 (rank : 9) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91ZW3, Q8C791, Q8CA22, Q8VDG1, Q925M9 | Gene names | Smarca5, Snf2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (Sucrose nonfermenting protein 2 homolog) (mSnf2h). | |||||
|
SMCA5_HUMAN
|
||||||
| NC score | 0.860368 (rank : 10) | θ value | 9.20422e-37 (rank : 10) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60264 | Gene names | SMARCA5, SNF2H, WCRF135 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin A5) (Sucrose nonfermenting protein 2 homolog) (hSNF2H). | |||||
|
SMCA1_MOUSE
|
||||||
| NC score | 0.859726 (rank : 11) | θ value | 2.34062e-40 (rank : 7) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
ERCC6_HUMAN
|
||||||
| NC score | 0.859580 (rank : 12) | θ value | 1.32909e-35 (rank : 11) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
RAD54_HUMAN
|
||||||
| NC score | 0.858056 (rank : 13) | θ value | 8.93572e-24 (rank : 24) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92698, Q6IUY3 | Gene names | RAD54L, RAD54A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (hRAD54) (hHR54). | |||||
|
RAD54_MOUSE
|
||||||
| NC score | 0.854351 (rank : 14) | θ value | 8.36355e-22 (rank : 27) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70270, Q8BSR5, Q8C2C4, Q8K3D4 | Gene names | Rad54l, Rad54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (mRAD54) (mHR54). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.831350 (rank : 15) | θ value | 5.22648e-32 (rank : 14) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.829498 (rank : 16) | θ value | 2.86786e-30 (rank : 16) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
INOC1_HUMAN
|
||||||
| NC score | 0.828505 (rank : 17) | θ value | 1.5242e-23 (rank : 25) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
INOC1_MOUSE
|
||||||
| NC score | 0.827995 (rank : 18) | θ value | 3.39556e-23 (rank : 26) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6ZPV2, Q6P7V0, Q6PCP1, Q8C9T7 | Gene names | Inoc1, Kiaa1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.827532 (rank : 19) | θ value | 2.42779e-29 (rank : 21) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.824725 (rank : 20) | θ value | 3.74554e-30 (rank : 17) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.824640 (rank : 21) | θ value | 2.42779e-29 (rank : 20) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.818897 (rank : 22) | θ value | 6.38894e-30 (rank : 19) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.814292 (rank : 23) | θ value | 3.74554e-30 (rank : 18) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
SMRA3_MOUSE
|
||||||
| NC score | 0.813398 (rank : 24) | θ value | 4.84666e-62 (rank : 4) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6PCN7, O35596, O35597, Q80VT6 | Gene names | Smarca3, Snf2l3, Zbu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (TNF-response element-binding protein) (P113). | |||||
|
SMRA3_HUMAN
|
||||||
| NC score | 0.809813 (rank : 25) | θ value | 2.24614e-67 (rank : 3) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14527, Q14536, Q16051, Q7KYJ6, Q86YA5, Q92652, Q96KM9 | Gene names | SMARCA3, HIP116A, HLTF, SNF2L3, ZBU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (DNA-binding protein/plasminogen activator inhibitor 1 regulator) (Helicase-like transcription factor) (HIP116). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.805813 (rank : 26) | θ value | 4.14116e-29 (rank : 22) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.792921 (rank : 27) | θ value | 6.61148e-27 (rank : 23) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SMAL1_MOUSE
|
||||||
| NC score | 0.771264 (rank : 28) | θ value | 4.16044e-13 (rank : 37) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.765400 (rank : 29) | θ value | 1.42992e-13 (rank : 36) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.746341 (rank : 30) | θ value | 3.28887e-18 (rank : 28) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.745229 (rank : 31) | θ value | 8.10077e-17 (rank : 31) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.744549 (rank : 32) | θ value | 5.60996e-18 (rank : 30) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.743306 (rank : 33) | θ value | 3.28887e-18 (rank : 29) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.728147 (rank : 34) | θ value | 1.68911e-14 (rank : 35) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.722482 (rank : 35) | θ value | 1.16975e-15 (rank : 33) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.661297 (rank : 36) | θ value | 5.25075e-16 (rank : 32) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.641503 (rank : 37) | θ value | 2.60593e-15 (rank : 34) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
NEIL3_HUMAN
|
||||||
| NC score | 0.105243 (rank : 38) | θ value | 0.00298849 (rank : 38) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
NEIL3_MOUSE
|
||||||
| NC score | 0.098608 (rank : 39) | θ value | 0.00869519 (rank : 40) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.067338 (rank : 40) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.067162 (rank : 41) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.065254 (rank : 42) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.064821 (rank : 43) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.064414 (rank : 44) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.064091 (rank : 45) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.061902 (rank : 46) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.061535 (rank : 47) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.060704 (rank : 48) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CBX5_HUMAN
|
||||||
| NC score | 0.060603 (rank : 49) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.059083 (rank : 50) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.058788 (rank : 51) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.058261 (rank : 52) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.057361 (rank : 53) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.056912 (rank : 54) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.056662 (rank : 55) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.056579 (rank : 56) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.056487 (rank : 57) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.056257 (rank : 58) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.055267 (rank : 59) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.054504 (rank : 60) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.054349 (rank : 61) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.054103 (rank : 62) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.054027 (rank : 63) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.053769 (rank : 64) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
CBX5_MOUSE
|
||||||
| NC score | 0.053284 (rank : 65) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX1_HUMAN
|
||||||
| NC score | 0.052074 (rank : 66) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| NC score | 0.052074 (rank : 67) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
SP110_HUMAN
|
||||||
| NC score | 0.051319 (rank : 68) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.051178 (rank : 69) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.050532 (rank : 70) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
NPDC1_MOUSE
|
||||||
| NC score | 0.044240 (rank : 71) | θ value | 0.0736092 (rank : 44) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64322 | Gene names | Npdc1, Npdc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Neural proliferation differentiation and control protein 1 precursor (NPDC-1 protein). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.038094 (rank : 72) | θ value | 0.00869519 (rank : 39) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.037707 (rank : 73) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
TOP3A_HUMAN
|
||||||
| NC score | 0.037408 (rank : 74) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13472, Q13473 | Gene names | TOP3A, TOP3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.036423 (rank : 75) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
APEX2_HUMAN
|
||||||
| NC score | 0.035409 (rank : 76) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UBZ4, Q9Y5X7 | Gene names | APEX2, APE2, APEXL2, XTH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2) (APEX nuclease-like 2) (AP endonuclease XTH2). | |||||
|
TOP3A_MOUSE
|
||||||
| NC score | 0.034838 (rank : 77) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70157 | Gene names | Top3a, Top3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.033387 (rank : 78) | θ value | 0.0563607 (rank : 43) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
APEX2_MOUSE
|
||||||
| NC score | 0.033234 (rank : 79) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68G58, Q8BJP7, Q8BTR7, Q8BUZ2, Q8BYE9, Q8R018, Q8R328, Q9CS12 | Gene names | Apex2, Ape2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.030030 (rank : 80) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.028496 (rank : 81) | θ value | 0.0736092 (rank : 45) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.027580 (rank : 82) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
MGAT5_HUMAN
|
||||||
| NC score | 0.026461 (rank : 83) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q09328 | Gene names | MGAT5, GGNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V (EC 2.4.1.155) (Mannoside acetylglucosaminyltransferase 5) (Alpha- mannoside beta-1,6-N-acetylglucosaminyltransferase) (N- acetylglucosaminyl-transferase V) (GNT-V) (GlcNAc-T V). | |||||
|
MGAT5_MOUSE
|
||||||
| NC score | 0.026441 (rank : 84) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4G6 | Gene names | Mgat5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V (EC 2.4.1.155) (Mannoside acetylglucosaminyltransferase 5) (Alpha- mannoside beta-1,6-N-acetylglucosaminyltransferase) (N- acetylglucosaminyl-transferase V) (GNT-V) (GlcNAc-T V). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.023045 (rank : 85) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.022591 (rank : 86) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.020940 (rank : 87) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
TRI29_HUMAN
|
||||||
| NC score | 0.020261 (rank : 88) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14134, Q96AA9, Q9BZY7 | Gene names | TRIM29, ATDC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 29 (Ataxia-telangiectasia group D- associated protein). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.020120 (rank : 89) | θ value | 0.0330416 (rank : 41) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
ZN451_HUMAN
|
||||||
| NC score | 0.019859 (rank : 90) | θ value | 0.0330416 (rank : 42) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 627 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4E5, Q5VVF1, Q8N380, Q8TD15, Q9NQM1 | Gene names | ZNF451, COASTER, KIAA0576 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 451 (Coactivator for steroid receptors). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.019487 (rank : 91) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
EFHC2_HUMAN
|
||||||
| NC score | 0.019355 (rank : 92) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JST6, Q5JST8, Q68DK4, Q8NEI0, Q9H653 | Gene names | EFHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member C2. | |||||
|
NFAT5_MOUSE
|
||||||
| NC score | 0.018519 (rank : 93) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
CNGB3_MOUSE
|
||||||
| NC score | 0.017910 (rank : 94) | θ value | 0.0961366 (rank : 46) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.016356 (rank : 95) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
DPOLA_MOUSE
|
||||||
| NC score | 0.016168 (rank : 96) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33609 | Gene names | Pola1, Pola | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.015796 (rank : 97) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CF064_MOUSE
|
||||||
| NC score | 0.015482 (rank : 98) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K190 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf64 homolog. | |||||
|
ZN451_MOUSE
|
||||||
| NC score | 0.015454 (rank : 99) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0P7 | Gene names | Znf451, Zfp451 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 451. | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.014908 (rank : 100) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.013824 (rank : 101) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
ZW10_HUMAN
|
||||||
| NC score | 0.013012 (rank : 102) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43264 | Gene names | ZW10 | |||
|
Domain Architecture |

|
|||||
| Description | Centromere/kinetochore protein zw10 homolog. | |||||
|
HASP_MOUSE
|
||||||
| NC score | 0.012699 (rank : 103) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0R0, Q99MU9, Q9JJJ4 | Gene names | Gsg2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Haspin (EC 2.7.11.1) (Haploid germ cell-specific nuclear protein kinase) (Germ cell-specific gene 2 protein). | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.012611 (rank : 104) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
DPOLA_HUMAN
|
||||||
| NC score | 0.012480 (rank : 105) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09884, Q86UQ7 | Gene names | POLA1, POLA | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
|
WDHD1_MOUSE
|
||||||
| NC score | 0.012439 (rank : 106) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59328, Q6P408 | Gene names | Wdhd1, And1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.012337 (rank : 107) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
TCEA1_HUMAN
|
||||||
| NC score | 0.011946 (rank : 108) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23193, Q15563, Q6FG87 | Gene names | TCEA1, TFIIS | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o). | |||||
|
STX12_HUMAN
|
||||||
| NC score | 0.011832 (rank : 109) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86Y82, O95564 | Gene names | STX12 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-12. | |||||
|
SIA7A_MOUSE
|
||||||
| NC score | 0.011537 (rank : 110) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZ39, Q9JJP5 | Gene names | St6galnac1, Siat7a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
STX12_MOUSE
|
||||||
| NC score | 0.011063 (rank : 111) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ER00, Q3UIV9, Q921T9 | Gene names | Stx12 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-12. | |||||
|
ATF1_MOUSE
|
||||||
| NC score | 0.011059 (rank : 112) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
SPC25_MOUSE
|
||||||
| NC score | 0.011005 (rank : 113) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UA16, Q9D021, Q9D1K6 | Gene names | Spbc25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25. | |||||
|
EZRI_HUMAN
|
||||||
| NC score | 0.009924 (rank : 114) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
DHX34_MOUSE
|
||||||
| NC score | 0.008578 (rank : 115) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.008093 (rank : 116) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
TUFT1_MOUSE
|
||||||
| NC score | 0.007282 (rank : 117) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.006460 (rank : 118) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.006453 (rank : 119) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.006053 (rank : 120) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
DEN2A_MOUSE
|
||||||
| NC score | 0.005566 (rank : 121) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C4S8, Q3TJU2, Q3TUG3, Q3UXA3 | Gene names | Dennd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.001297 (rank : 122) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
POT14_HUMAN
|
||||||
| NC score | 0.000992 (rank : 123) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6S5H5, Q6S5H6, Q6S8J2 | Gene names | POTE14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 14. | |||||
|
SNRK_MOUSE
|
||||||
| NC score | 0.000017 (rank : 124) | θ value | 0.21417 (rank : 47) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDU5, Q91WX6 | Gene names | Snrk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SNF-related serine/threonine-protein kinase (EC 2.7.11.1) (SNF1- related kinase). | |||||
|
SNRK_HUMAN
|
||||||
| NC score | -0.000377 (rank : 125) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRH2, Q14706, Q68D15, Q6IQ46, Q9NXI7 | Gene names | SNRK, KIAA0096, SNFRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SNF-related serine/threonine-protein kinase (EC 2.7.11.1) (SNF1- related kinase). | |||||
|
ZBT40_HUMAN
|
||||||
| NC score | -0.000720 (rank : 126) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
ZN690_HUMAN
|
||||||
| NC score | -0.003547 (rank : 127) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IWY8, Q32M75, Q32M76, Q8NA40 | Gene names | ZNF690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 690. | |||||
|
O2T33_HUMAN
|
||||||
| NC score | -0.004220 (rank : 128) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NG76 | Gene names | OR2T33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 2T33 (Olfactory receptor OR1-56). | |||||