Please be patient as the page loads
|
BTAF1_HUMAN
|
||||||
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
BTAF1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
ERCC6_HUMAN
|
||||||
| θ value | 4.36332e-79 (rank : 2) | NC score | 0.944390 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 3.4653e-68 (rank : 3) | NC score | 0.892848 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SMCA1_MOUSE
|
||||||
| θ value | 6.53529e-67 (rank : 4) | NC score | 0.938595 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
SMCA5_MOUSE
|
||||||
| θ value | 1.11475e-66 (rank : 5) | NC score | 0.942353 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91ZW3, Q8C791, Q8CA22, Q8VDG1, Q925M9 | Gene names | Smarca5, Snf2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (Sucrose nonfermenting protein 2 homolog) (mSnf2h). | |||||
|
SMCA5_HUMAN
|
||||||
| θ value | 2.4834e-66 (rank : 6) | NC score | 0.942275 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O60264 | Gene names | SMARCA5, SNF2H, WCRF135 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin A5) (Sucrose nonfermenting protein 2 homolog) (hSNF2H). | |||||
|
SMCA1_HUMAN
|
||||||
| θ value | 6.11685e-65 (rank : 7) | NC score | 0.940822 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P28370, Q5JV41, Q5JV42 | Gene names | SMARCA1, SNF2L, SNF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 2.56992e-63 (rank : 8) | NC score | 0.879239 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 4.38363e-63 (rank : 9) | NC score | 0.911676 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
SMRCD_HUMAN
|
||||||
| θ value | 2.17557e-62 (rank : 10) | NC score | 0.944614 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 4.84666e-62 (rank : 11) | NC score | 0.915564 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 6.99855e-61 (rank : 12) | NC score | 0.915704 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
RA54B_HUMAN
|
||||||
| θ value | 1.01059e-59 (rank : 13) | NC score | 0.942470 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y620 | Gene names | RAD54B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
SMRCD_MOUSE
|
||||||
| θ value | 2.7521e-57 (rank : 14) | NC score | 0.942626 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q04692 | Gene names | Smarcad1, Etl1, Kiaa1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase SMARCAD1) (Enhancer trap locus homolog 1) (Etl-1). | |||||
|
RAD54_HUMAN
|
||||||
| θ value | 4.69435e-57 (rank : 15) | NC score | 0.939818 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q92698, Q6IUY3 | Gene names | RAD54L, RAD54A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (hRAD54) (hHR54). | |||||
|
RAD54_MOUSE
|
||||||
| θ value | 1.78386e-56 (rank : 16) | NC score | 0.938409 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70270, Q8BSR5, Q8C2C4, Q8K3D4 | Gene names | Rad54l, Rad54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (mRAD54) (mHR54). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 6.77864e-56 (rank : 17) | NC score | 0.908691 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 7.49467e-55 (rank : 18) | NC score | 0.915195 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
RA54B_MOUSE
|
||||||
| θ value | 2.18062e-54 (rank : 19) | NC score | 0.939499 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6PFE3 | Gene names | Rad54b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 1.41344e-53 (rank : 20) | NC score | 0.903061 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 4.11246e-53 (rank : 21) | NC score | 0.898366 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 8.30557e-46 (rank : 22) | NC score | 0.840771 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 8.30557e-46 (rank : 23) | NC score | 0.837271 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 7.77379e-44 (rank : 24) | NC score | 0.839759 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 2.26182e-43 (rank : 25) | NC score | 0.841811 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
INOC1_HUMAN
|
||||||
| θ value | 8.04462e-41 (rank : 26) | NC score | 0.899470 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
INOC1_MOUSE
|
||||||
| θ value | 1.05066e-40 (rank : 27) | NC score | 0.899749 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6ZPV2, Q6P7V0, Q6PCP1, Q8C9T7 | Gene names | Inoc1, Kiaa1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-). | |||||
|
TTF2_HUMAN
|
||||||
| θ value | 1.12514e-34 (rank : 28) | NC score | 0.873290 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 7.06379e-29 (rank : 29) | NC score | 0.825131 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | 5.97985e-28 (rank : 30) | NC score | 0.840225 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
SMAL1_MOUSE
|
||||||
| θ value | 7.80994e-28 (rank : 31) | NC score | 0.849805 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 5.06226e-27 (rank : 32) | NC score | 0.805152 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 2.12685e-25 (rank : 33) | NC score | 0.796713 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
SMRA3_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 34) | NC score | 0.738742 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6PCN7, O35596, O35597, Q80VT6 | Gene names | Smarca3, Snf2l3, Zbu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (TNF-response element-binding protein) (P113). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 7.32683e-18 (rank : 35) | NC score | 0.692981 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
SMRA3_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 36) | NC score | 0.734482 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14527, Q14536, Q16051, Q7KYJ6, Q86YA5, Q92652, Q96KM9 | Gene names | SMARCA3, HIP116A, HLTF, SNF2L3, ZBU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (DNA-binding protein/plasminogen activator inhibitor 1 regulator) (Helicase-like transcription factor) (HIP116). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 37) | NC score | 0.673130 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 38) | NC score | 0.064693 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
DDX28_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 39) | NC score | 0.041483 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 0.125558 (rank : 40) | NC score | 0.072691 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
TNPO3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 41) | NC score | 0.025016 (rank : 102) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P2B1, Q7TSL6, Q8BKX4, Q8BP42 | Gene names | Tnpo3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transportin-3. | |||||
|
BMS1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.033447 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14692, Q86XJ9 | Gene names | BMS1L, KIAA0187 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BMS1 homolog. | |||||
|
DD19A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.030700 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.030730 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.030645 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 46) | NC score | 0.003354 (rank : 140) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
TNPO2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.035835 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14787, O14655 | Gene names | TNPO2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
TNPO2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.035904 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99LG2, Q8C0U9 | Gene names | Tnpo2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.030656 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
K0423_MOUSE
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.027958 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.063592 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
GRP75_HUMAN
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.010106 (rank : 123) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
K0423_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.025049 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
SYCP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.040737 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
CTGE6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.004644 (rank : 137) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86UF2 | Gene names | CTAGE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-6 (cTAGE family member 6). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.027096 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
TNPO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.032968 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92973, Q92957, Q92975 | Gene names | TNPO1, KPNB2, MIP1, TRN | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2) (M9 region interaction protein) (MIP). | |||||
|
TNPO1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.033003 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BFY9, Q8C0S3, Q8K2E7 | Gene names | Tnpo1, Kpnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2). | |||||
|
VPS54_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.015724 (rank : 111) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P1Q0, Q5VIR5, Q86YF7, Q8N6G3, Q9NPV0, Q9NT07, Q9NUJ0 | Gene names | VPS54, HCC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 54 (Tumor antigen SLP-8p) (Hepatocellular carcinoma protein 8). | |||||
|
ZC3H8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.017846 (rank : 106) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
BAG3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.016501 (rank : 110) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
IMB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.024337 (rank : 103) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14974, Q14637, Q96J27 | Gene names | KPNB1, NTF97 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Importin 90). | |||||
|
RNBP6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.017025 (rank : 107) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60518, Q5T7X4, Q7Z3V2, Q96E78 | Gene names | RANBP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 6 (RanBP6). | |||||
|
TLK2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | -0.001900 (rank : 143) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55047, Q9D5Y5 | Gene names | Tlk2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2). | |||||
|
GCN1L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.018256 (rank : 105) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92616, O95001, O95651, Q6P2S3, Q86X65, Q8N5I5, Q8WU80, Q99736, Q9UE60 | Gene names | GCN1L1, KIAA0219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GCN1-like protein 1 (HsGCN1). | |||||
|
GRP75_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.008345 (rank : 126) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
DNMT1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.027913 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
JHD2B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.006174 (rank : 134) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
DMAP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.015303 (rank : 113) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPF5, Q5TG41, Q7Z3H7, Q9H0S8, Q9P2C2 | Gene names | DMAP1, KIAA1425 | |||
|
Domain Architecture |

|
|||||
| Description | DNA methyltransferase 1-associated protein 1 (DNMT1-associated protein 1) (DNMAP1). | |||||
|
DMAP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.015319 (rank : 112) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JI44, Q99LM0, Q9CSS9, Q9DB33 | Gene names | Dmap1, Mmtr | |||
|
Domain Architecture |

|
|||||
| Description | DNA methyltransferase 1-associated protein 1 (DNMT1-associated protein 1) (DNMAP1) (MAT1-mediated transcriptional repressor). | |||||
|
HD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.008257 (rank : 127) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
K1H1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | -0.000265 (rank : 141) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15323 | Gene names | KRTHA1, HHA1, HKA1 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cuticular Ha1 (Hair keratin, type I Ha1). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.012061 (rank : 118) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
UTP20_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.013196 (rank : 115) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75691, Q9H3H4 | Gene names | UTP20, DRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein) (Protein Key-1A6) (Novel nucleolar protein 73) (NNP73). | |||||
|
UTP20_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.013167 (rank : 116) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5XG71, Q80V22, Q8BXH9, Q8CHL6, Q99K11 | Gene names | Utp20, Drim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein). | |||||
|
XPO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.016575 (rank : 109) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14980, Q63HP8, Q68CP3, Q99433 | Gene names | XPO1, CRM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-1 (Exp1) (Chromosome region maintenance 1 protein homolog). | |||||
|
XPO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.016872 (rank : 108) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P5F9 | Gene names | Xpo1, Crm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-1 (Exp1) (Chromosome region maintenance 1 protein homolog). | |||||
|
2AAA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.011135 (rank : 122) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P30153, Q13773 | Gene names | PPP2R1A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A, subunit A, PR65-alpha isoform) (PP2A, subunit A, R1-alpha isoform) (Medium tumor antigen-associated 61 kDa protein). | |||||
|
2AAA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.011416 (rank : 120) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q76MZ3 | Gene names | Ppp2r1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A, subunit A, PR65-alpha isoform) (PP2A, subunit A, R1-alpha isoform). | |||||
|
ACY1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.013234 (rank : 114) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03154 | Gene names | ACY1 | |||
|
Domain Architecture |

|
|||||
| Description | Aminoacylase-1 (EC 3.5.1.14) (N-acyl-L-amino-acid amidohydrolase) (ACY-1). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.004104 (rank : 138) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.006109 (rank : 135) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
GOSR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.010051 (rank : 124) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95249, O75392 | Gene names | GOSR1, GS28 | |||
|
Domain Architecture |
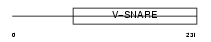
|
|||||
| Description | Golgi SNAP receptor complex member 1 (28 kDa Golgi SNARE protein) (28 kDa cis-Golgi SNARE p28) (GOS-28). | |||||
|
IFT20_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.012999 (rank : 117) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IY31, Q5GLZ2, Q9BUG5 | Gene names | IFT20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 20 homolog (Intraflagellar transport protein 20) (hIFT20). | |||||
|
IPO11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.011403 (rank : 121) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K2V6, Q8BU45, Q8K0B4 | Gene names | Ipo11, Ranbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-11 (Imp11) (Ran-binding protein 11) (RanBP11). | |||||
|
RU2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.006727 (rank : 132) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08579, Q9UJD4 | Gene names | SNRPB2 | |||
|
Domain Architecture |
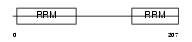
|
|||||
| Description | U2 small nuclear ribonucleoprotein B''. | |||||
|
TAD3L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.007264 (rank : 131) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0L9, Q8CIH4, Q8K289, Q8R5E3, Q9CTJ0 | Gene names | Tada3l, Ada3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional adapter 3-like (ADA3-like protein) (ADA3 homolog) (mADA3). | |||||
|
WFDC8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.007512 (rank : 129) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
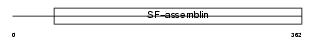
Domain Architecture |
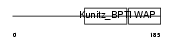
|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
ZC3H8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.011514 (rank : 119) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.024015 (rank : 104) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
CI072_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.007443 (rank : 130) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6DFW0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf72 homolog. | |||||
|
DBF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.006720 (rank : 133) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBU7, O75226, Q75MS6, Q75N01, Q9Y2M6 | Gene names | DBF4, ASK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DBF4 homolog (Activator of S phase Kinase). | |||||
|
ECM29_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.009697 (rank : 125) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PDI5, Q6ZQC9, Q8BSW7, Q8CAH0, Q8R3M6 | Gene names | Ecm29, Kiaa0368 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome-associated protein ECM29 homolog (Ecm29). | |||||
|
STX7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.003966 (rank : 139) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15400, Q5SZW2, Q96ES9 | Gene names | STX7 | |||
|
Domain Architecture |
|
|||||
| Description | Syntaxin-7. | |||||
|
TIP60_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.004763 (rank : 136) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHK4, Q8CGZ3, Q8CGZ4, Q8VIH0 | Gene names | Htatip, Tip60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60). | |||||
|
TLK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | -0.001213 (rank : 142) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UE8, Q9UKI7, Q9Y4F7 | Gene names | TLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2) (PKU-alpha). | |||||
|
XPO5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.008066 (rank : 128) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HAV4, Q5JTE6, Q96G48, Q96HN3, Q9BWM6, Q9BZV5, Q9H9M4, Q9NT89, Q9NW39, Q9ULC9 | Gene names | XPO5, KIAA1291, RANBP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-5 (Exp5) (Ran-binding protein 21). | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.085126 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.086417 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.063434 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.060653 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.068660 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.081972 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CBX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.063452 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.063452 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.053983 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.056985 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CBX5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.075864 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.067519 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.055431 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.058837 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.061433 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.066270 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.058782 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.061270 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LY10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.059234 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.059409 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.054483 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.051869 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.054748 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.054890 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.072352 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.073127 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.074617 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.075138 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.080697 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.080687 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.077731 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.051949 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.052596 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.065318 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SP110_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.066185 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.050544 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.066085 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TIF1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.059436 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.062356 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.055023 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.061794 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.062023 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRI66_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.061313 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.051590 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.072853 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.070685 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
BTAF1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
SMRCD_HUMAN
|
||||||
| NC score | 0.944614 (rank : 2) | θ value | 2.17557e-62 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
|
ERCC6_HUMAN
|
||||||
| NC score | 0.944390 (rank : 3) | θ value | 4.36332e-79 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
SMRCD_MOUSE
|
||||||
| NC score | 0.942626 (rank : 4) | θ value | 2.7521e-57 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q04692 | Gene names | Smarcad1, Etl1, Kiaa1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase SMARCAD1) (Enhancer trap locus homolog 1) (Etl-1). | |||||
|
RA54B_HUMAN
|
||||||
| NC score | 0.942470 (rank : 5) | θ value | 1.01059e-59 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y620 | Gene names | RAD54B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
SMCA5_MOUSE
|
||||||
| NC score | 0.942353 (rank : 6) | θ value | 1.11475e-66 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91ZW3, Q8C791, Q8CA22, Q8VDG1, Q925M9 | Gene names | Smarca5, Snf2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (Sucrose nonfermenting protein 2 homolog) (mSnf2h). | |||||
|
SMCA5_HUMAN
|
||||||
| NC score | 0.942275 (rank : 7) | θ value | 2.4834e-66 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O60264 | Gene names | SMARCA5, SNF2H, WCRF135 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin A5) (Sucrose nonfermenting protein 2 homolog) (hSNF2H). | |||||
|
SMCA1_HUMAN
|
||||||
| NC score | 0.940822 (rank : 8) | θ value | 6.11685e-65 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P28370, Q5JV41, Q5JV42 | Gene names | SMARCA1, SNF2L, SNF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1). | |||||
|
RAD54_HUMAN
|
||||||
| NC score | 0.939818 (rank : 9) | θ value | 4.69435e-57 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q92698, Q6IUY3 | Gene names | RAD54L, RAD54A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (hRAD54) (hHR54). | |||||
|
RA54B_MOUSE
|
||||||
| NC score | 0.939499 (rank : 10) | θ value | 2.18062e-54 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6PFE3 | Gene names | Rad54b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
SMCA1_MOUSE
|
||||||
| NC score | 0.938595 (rank : 11) | θ value | 6.53529e-67 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
RAD54_MOUSE
|
||||||
| NC score | 0.938409 (rank : 12) | θ value | 1.78386e-56 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70270, Q8BSR5, Q8C2C4, Q8K3D4 | Gene names | Rad54l, Rad54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (mRAD54) (mHR54). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.915704 (rank : 13) | θ value | 6.99855e-61 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.915564 (rank : 14) | θ value | 4.84666e-62 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.915195 (rank : 15) | θ value | 7.49467e-55 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.911676 (rank : 16) | θ value | 4.38363e-63 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.908691 (rank : 17) | θ value | 6.77864e-56 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.903061 (rank : 18) | θ value | 1.41344e-53 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
INOC1_MOUSE
|
||||||
| NC score | 0.899749 (rank : 19) | θ value | 1.05066e-40 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6ZPV2, Q6P7V0, Q6PCP1, Q8C9T7 | Gene names | Inoc1, Kiaa1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-). | |||||
|
INOC1_HUMAN
|
||||||
| NC score | 0.899470 (rank : 20) | θ value | 8.04462e-41 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.898366 (rank : 21) | θ value | 4.11246e-53 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.892848 (rank : 22) | θ value | 3.4653e-68 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.879239 (rank : 23) | θ value | 2.56992e-63 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
TTF2_HUMAN
|
||||||
| NC score | 0.873290 (rank : 24) | θ value | 1.12514e-34 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
SMAL1_MOUSE
|
||||||
| NC score | 0.849805 (rank : 25) | θ value | 7.80994e-28 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.841811 (rank : 26) | θ value | 2.26182e-43 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.840771 (rank : 27) | θ value | 8.30557e-46 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.840225 (rank : 28) | θ value | 5.97985e-28 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.839759 (rank : 29) | θ value | 7.77379e-44 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.837271 (rank : 30) | θ value | 8.30557e-46 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.825131 (rank : 31) | θ value | 7.06379e-29 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.805152 (rank : 32) | θ value | 5.06226e-27 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.796713 (rank : 33) | θ value | 2.12685e-25 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
SMRA3_MOUSE
|
||||||
| NC score | 0.738742 (rank : 34) | θ value | 3.88503e-19 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6PCN7, O35596, O35597, Q80VT6 | Gene names | Smarca3, Snf2l3, Zbu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (TNF-response element-binding protein) (P113). | |||||
|
SMRA3_HUMAN
|
||||||
| NC score | 0.734482 (rank : 35) | θ value | 7.32683e-18 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14527, Q14536, Q16051, Q7KYJ6, Q86YA5, Q92652, Q96KM9 | Gene names | SMARCA3, HIP116A, HLTF, SNF2L3, ZBU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (DNA-binding protein/plasminogen activator inhibitor 1 regulator) (Helicase-like transcription factor) (HIP116). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.692981 (rank : 36) | θ value | 7.32683e-18 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.673130 (rank : 37) | θ value | 1.63225e-17 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.086417 (rank : 38) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.085126 (rank : 39) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.081972 (rank : 40) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.080697 (rank : 41) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.080687 (rank : 42) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.077731 (rank : 43) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
CBX5_HUMAN
|
||||||
| NC score | 0.075864 (rank : 44) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.075138 (rank : 45) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.074617 (rank : 46) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.073127 (rank : 47) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.072853 (rank : 48) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.072691 (rank : 49) | θ value | 0.125558 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.072352 (rank : 50) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.070685 (rank : 51) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.068660 (rank : 52) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
CBX5_MOUSE
|
||||||
| NC score | 0.067519 (rank : 53) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.066270 (rank : 54) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SP110_HUMAN
|
||||||
| NC score | 0.066185 (rank : 55) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.066085 (rank : 56) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.065318 (rank : 57) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.064693 (rank : 58) | θ value | 0.0252991 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.063592 (rank : 59) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
CBX1_HUMAN
|
||||||
| NC score | 0.063452 (rank : 60) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| NC score | 0.063452 (rank : 61) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.063434 (rank : 62) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.062356 (rank : 63) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.062023 (rank : 64) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.061794 (rank : 65) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.061433 (rank : 66) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.061313 (rank : 67) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.061270 (rank : 68) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.060653 (rank : 69) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
TIF1A_HUMAN
|
||||||
| NC score | 0.059436 (rank : 70) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.059409 (rank : 71) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
LY10_HUMAN
|
||||||
| NC score | 0.059234 (rank : 72) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
CBX7_MOUSE
|
||||||
| NC score | 0.058837 (rank : 73) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.058782 (rank : 74) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
CBX3_MOUSE
|
||||||
| NC score | 0.056985 (rank : 75) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CBX7_HUMAN
|
||||||
| NC score | 0.055431 (rank : 76) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.055023 (rank : 77) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.054890 (rank : 78) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.054748 (rank : 79) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.054483 (rank : 80) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CBX3_HUMAN
|
||||||
| NC score | 0.053983 (rank : 81) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.052596 (rank : 82) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.051949 (rank : 83) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.051869 (rank : 84) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.051590 (rank : 85) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.050544 (rank : 86) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.041483 (rank : 87) | θ value | 0.0961366 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
SYCP2_HUMAN
|
||||||
| NC score | 0.040737 (rank : 88) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
TNPO2_MOUSE
|
||||||
| NC score | 0.035904 (rank : 89) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99LG2, Q8C0U9 | Gene names | Tnpo2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
TNPO2_HUMAN
|
||||||
| NC score | 0.035835 (rank : 90) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14787, O14655 | Gene names | TNPO2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
BMS1_HUMAN
|
||||||
| NC score | 0.033447 (rank : 91) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14692, Q86XJ9 | Gene names | BMS1L, KIAA0187 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BMS1 homolog. | |||||
|
TNPO1_MOUSE
|
||||||
| NC score | 0.033003 (rank : 92) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BFY9, Q8C0S3, Q8K2E7 | Gene names | Tnpo1, Kpnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2). | |||||
|
TNPO1_HUMAN
|
||||||
| NC score | 0.032968 (rank : 93) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92973, Q92957, Q92975 | Gene names | TNPO1, KPNB2, MIP1, TRN | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2) (M9 region interaction protein) (MIP). | |||||
|
DD19A_MOUSE
|
||||||
| NC score | 0.030730 (rank : 94) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19A_HUMAN
|
||||||
| NC score | 0.030700 (rank : 95) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.030656 (rank : 96) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
DD19B_HUMAN
|
||||||
| NC score | 0.030645 (rank : 97) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
K0423_MOUSE
|
||||||
| NC score | 0.027958 (rank : 98) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
DNMT1_HUMAN
|
||||||
| NC score | 0.027913 (rank : 99) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.027096 (rank : 100) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
K0423_HUMAN
|
||||||
| NC score | 0.025049 (rank : 101) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
TNPO3_MOUSE
|
||||||
| NC score | 0.025016 (rank : 102) | θ value | 0.21417 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P2B1, Q7TSL6, Q8BKX4, Q8BP42 | Gene names | Tnpo3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transportin-3. | |||||
|
IMB1_HUMAN
|
||||||
| NC score | 0.024337 (rank : 103) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14974, Q14637, Q96J27 | Gene names | KPNB1, NTF97 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Importin 90). | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.024015 (rank : 104) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
GCN1L_HUMAN
|
||||||
| NC score | 0.018256 (rank : 105) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92616, O95001, O95651, Q6P2S3, Q86X65, Q8N5I5, Q8WU80, Q99736, Q9UE60 | Gene names | GCN1L1, KIAA0219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GCN1-like protein 1 (HsGCN1). | |||||
|
ZC3H8_HUMAN
|
||||||
| NC score | 0.017846 (rank : 106) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
RNBP6_HUMAN
|
||||||
| NC score | 0.017025 (rank : 107) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60518, Q5T7X4, Q7Z3V2, Q96E78 | Gene names | RANBP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 6 (RanBP6). | |||||
|
XPO1_MOUSE
|
||||||
| NC score | 0.016872 (rank : 108) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P5F9 | Gene names | Xpo1, Crm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-1 (Exp1) (Chromosome region maintenance 1 protein homolog). | |||||
|
XPO1_HUMAN
|
||||||
| NC score | 0.016575 (rank : 109) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14980, Q63HP8, Q68CP3, Q99433 | Gene names | XPO1, CRM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-1 (Exp1) (Chromosome region maintenance 1 protein homolog). | |||||
|
BAG3_HUMAN
|
||||||
| NC score | 0.016501 (rank : 110) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
VPS54_HUMAN
|
||||||
| NC score | 0.015724 (rank : 111) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P1Q0, Q5VIR5, Q86YF7, Q8N6G3, Q9NPV0, Q9NT07, Q9NUJ0 | Gene names | VPS54, HCC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 54 (Tumor antigen SLP-8p) (Hepatocellular carcinoma protein 8). | |||||
|
DMAP1_MOUSE
|
||||||
| NC score | 0.015319 (rank : 112) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JI44, Q99LM0, Q9CSS9, Q9DB33 | Gene names | Dmap1, Mmtr | |||
|
Domain Architecture |

|
|||||
| Description | DNA methyltransferase 1-associated protein 1 (DNMT1-associated protein 1) (DNMAP1) (MAT1-mediated transcriptional repressor). | |||||
|
DMAP1_HUMAN
|
||||||
| NC score | 0.015303 (rank : 113) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPF5, Q5TG41, Q7Z3H7, Q9H0S8, Q9P2C2 | Gene names | DMAP1, KIAA1425 | |||
|
Domain Architecture |

|
|||||
| Description | DNA methyltransferase 1-associated protein 1 (DNMT1-associated protein 1) (DNMAP1). | |||||
|
ACY1_HUMAN
|
||||||
| NC score | 0.013234 (rank : 114) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03154 | Gene names | ACY1 | |||
|
Domain Architecture |

|
|||||
| Description | Aminoacylase-1 (EC 3.5.1.14) (N-acyl-L-amino-acid amidohydrolase) (ACY-1). | |||||
|
UTP20_HUMAN
|
||||||
| NC score | 0.013196 (rank : 115) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75691, Q9H3H4 | Gene names | UTP20, DRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein) (Protein Key-1A6) (Novel nucleolar protein 73) (NNP73). | |||||
|
UTP20_MOUSE
|
||||||
| NC score | 0.013167 (rank : 116) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5XG71, Q80V22, Q8BXH9, Q8CHL6, Q99K11 | Gene names | Utp20, Drim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein). | |||||
|
IFT20_HUMAN
|
||||||
| NC score | 0.012999 (rank : 117) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IY31, Q5GLZ2, Q9BUG5 | Gene names | IFT20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 20 homolog (Intraflagellar transport protein 20) (hIFT20). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.012061 (rank : 118) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
ZC3H8_MOUSE
|
||||||
| NC score | 0.011514 (rank : 119) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
2AAA_MOUSE
|
||||||
| NC score | 0.011416 (rank : 120) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q76MZ3 | Gene names | Ppp2r1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A, subunit A, PR65-alpha isoform) (PP2A, subunit A, R1-alpha isoform). | |||||
|
IPO11_MOUSE
|
||||||
| NC score | 0.011403 (rank : 121) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K2V6, Q8BU45, Q8K0B4 | Gene names | Ipo11, Ranbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-11 (Imp11) (Ran-binding protein 11) (RanBP11). | |||||
|
2AAA_HUMAN
|
||||||
| NC score | 0.011135 (rank : 122) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P30153, Q13773 | Gene names | PPP2R1A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A, subunit A, PR65-alpha isoform) (PP2A, subunit A, R1-alpha isoform) (Medium tumor antigen-associated 61 kDa protein). | |||||
|
GRP75_HUMAN
|
||||||
| NC score | 0.010106 (rank : 123) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
GOSR1_HUMAN
|
||||||
| NC score | 0.010051 (rank : 124) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95249, O75392 | Gene names | GOSR1, GS28 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi SNAP receptor complex member 1 (28 kDa Golgi SNARE protein) (28 kDa cis-Golgi SNARE p28) (GOS-28). | |||||
|
ECM29_MOUSE
|
||||||
| NC score | 0.009697 (rank : 125) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PDI5, Q6ZQC9, Q8BSW7, Q8CAH0, Q8R3M6 | Gene names | Ecm29, Kiaa0368 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome-associated protein ECM29 homolog (Ecm29). | |||||
|
GRP75_MOUSE
|
||||||
| NC score | 0.008345 (rank : 126) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
HD_HUMAN
|
||||||
| NC score | 0.008257 (rank : 127) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
XPO5_HUMAN
|
||||||
| NC score | 0.008066 (rank : 128) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HAV4, Q5JTE6, Q96G48, Q96HN3, Q9BWM6, Q9BZV5, Q9H9M4, Q9NT89, Q9NW39, Q9ULC9 | Gene names | XPO5, KIAA1291, RANBP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-5 (Exp5) (Ran-binding protein 21). | |||||
|
WFDC8_HUMAN
|
||||||
| NC score | 0.007512 (rank : 129) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
CI072_MOUSE
|
||||||
| NC score | 0.007443 (rank : 130) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6DFW0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf72 homolog. | |||||
|
TAD3L_MOUSE
|
||||||
| NC score | 0.007264 (rank : 131) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0L9, Q8CIH4, Q8K289, Q8R5E3, Q9CTJ0 | Gene names | Tada3l, Ada3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional adapter 3-like (ADA3-like protein) (ADA3 homolog) (mADA3). | |||||
|
RU2B_HUMAN
|
||||||
| NC score | 0.006727 (rank : 132) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08579, Q9UJD4 | Gene names | SNRPB2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein B''. | |||||
|
DBF4_HUMAN
|
||||||
| NC score | 0.006720 (rank : 133) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBU7, O75226, Q75MS6, Q75N01, Q9Y2M6 | Gene names | DBF4, ASK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DBF4 homolog (Activator of S phase Kinase). | |||||
|
JHD2B_HUMAN
|
||||||
| NC score | 0.006174 (rank : 134) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.006109 (rank : 135) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
TIP60_MOUSE
|
||||||
| NC score | 0.004763 (rank : 136) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHK4, Q8CGZ3, Q8CGZ4, Q8VIH0 | Gene names | Htatip, Tip60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60). | |||||
|
CTGE6_HUMAN
|
||||||
| NC score | 0.004644 (rank : 137) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86UF2 | Gene names | CTAGE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-6 (cTAGE family member 6). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.004104 (rank : 138) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
STX7_HUMAN
|
||||||
| NC score | 0.003966 (rank : 139) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15400, Q5SZW2, Q96ES9 | Gene names | STX7 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-7. | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.003354 (rank : 140) | θ value | 0.47712 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
K1H1_HUMAN
|
||||||
| NC score | -0.000265 (rank : 141) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15323 | Gene names | KRTHA1, HHA1, HKA1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha1 (Hair keratin, type I Ha1). | |||||
|
TLK2_HUMAN
|
||||||
| NC score | -0.001213 (rank : 142) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UE8, Q9UKI7, Q9Y4F7 | Gene names | TLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2) (PKU-alpha). | |||||
|
TLK2_MOUSE
|
||||||
| NC score | -0.001900 (rank : 143) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55047, Q9D5Y5 | Gene names | Tlk2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2). | |||||