Please be patient as the page loads
|
WFDC8_HUMAN
|
||||||
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |
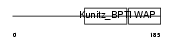
|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
WFDC8_HUMAN
|
||||||
| θ value | 1.05497e-141 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
EPPI_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 2) | NC score | 0.817635 (rank : 3) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95925, Q86TP9, Q96SD7, Q9HD30 | Gene names | SPINLW1, WAP7, WFDC7 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1) (WAP four-disulfide core domain protein 7) (Protease inhibitor WAP7). | |||||
|
EPPI_MOUSE
|
||||||
| θ value | 9.56915e-18 (rank : 3) | NC score | 0.827880 (rank : 2) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9DA01 | Gene names | Spinlw1 | |||
|
Domain Architecture |
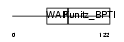
|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1). | |||||
|
WFDC3_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 4) | NC score | 0.466777 (rank : 20) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IUB2, Q8TC52, Q9BQP3, Q9BQP4 | Gene names | WFDC3, WAP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 3 precursor (Putative protease inhibitor WAP14). | |||||
|
WFDC5_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 5) | NC score | 0.543905 (rank : 13) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TCV5, Q6UWE4 | Gene names | WFDC5, WAP1 | |||
|
Domain Architecture |
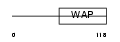
|
|||||
| Description | WAP four-disulfide core domain protein 5 precursor (Putative protease inhibitor WAP1). | |||||
|
WFD15_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 6) | NC score | 0.580176 (rank : 10) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JHY4, Q8BVC0 | Gene names | Wfdc15, Swam1 | |||
|
Domain Architecture |
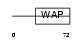
|
|||||
| Description | WAP four-disulfide core domain protein 15 precursor (Single WAP motif protein 1) (Elafin-like protein I). | |||||
|
WFDC6_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 7) | NC score | 0.662989 (rank : 4) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BQY6, Q5JYQ4, Q8NFV6 | Gene names | WFDC6, C20orf171, WAP6 | |||
|
Domain Architecture |
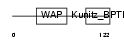
|
|||||
| Description | WAP four-disulfide core domain protein 6 precursor (Putative protease inhibitor WAP6). | |||||
|
TFPI2_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 8) | NC score | 0.612926 (rank : 7) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P48307 | Gene names | TFPI2 | |||
|
Domain Architecture |
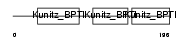
|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2) (Placental protein 5) (PP5). | |||||
|
TFPI2_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 9) | NC score | 0.613537 (rank : 6) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35536 | Gene names | Tfpi2 | |||
|
Domain Architecture |
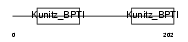
|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2). | |||||
|
AMBP_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 10) | NC score | 0.516207 (rank : 16) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P02760, P00977, P02759 | Gene names | AMBP, HCP, ITIL | |||
|
Domain Architecture |
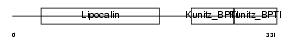
|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin (Protein HC) (Complex-forming glycoprotein heterogeneous in charge) (Alpha-1 microglycoprotein); Inter-alpha-trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
SPIT1_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 11) | NC score | 0.485813 (rank : 19) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9R097 | Gene names | Spint1, Hai1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
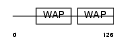
ALK1_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 12) | NC score | 0.528174 (rank : 14) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97430, O09081, O09082 | Gene names | Slpi | |||
|
Domain Architecture |
|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (Secretory leukocyte protease inhibitor). | |||||
|
A4_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 13) | NC score | 0.419639 (rank : 27) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 14) | NC score | 0.419530 (rank : 28) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
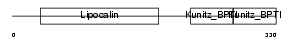
AMBP_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 15) | NC score | 0.499580 (rank : 18) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |
|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
APLP2_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 16) | NC score | 0.416018 (rank : 29) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
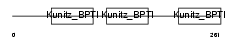
TFPI1_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 17) | NC score | 0.579843 (rank : 11) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P10646, O95103 | Gene names | TFPI, LACI, TFPI1 | |||
|
Domain Architecture |
|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
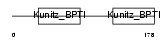
SPIT2_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 18) | NC score | 0.592163 (rank : 9) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43291, O00271, O14895, Q5TZQ3, Q969E0 | Gene names | SPINT2, HAI2, KOP | |||
|
Domain Architecture |
|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2) (Placental bikunin). | |||||
|
SPIT2_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 19) | NC score | 0.592829 (rank : 8) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WU03, Q9WU04, Q9WU05 | Gene names | Spint2, Hai2 | |||
|
Domain Architecture |
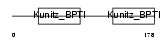
|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 20) | NC score | 0.118275 (rank : 43) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
WFD12_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 21) | NC score | 0.518496 (rank : 15) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWY7, Q5H980, Q9BR31 | Gene names | WFDC12, C20orf122, WAP2 | |||
|
Domain Architecture |
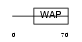
|
|||||
| Description | WAP four-disulfide core domain protein 12 precursor (Putative protease inhibitor WAP12) (Whey acidic protein 2). | |||||
|
WFDC2_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 22) | NC score | 0.448302 (rank : 23) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14508, Q8WXV9, Q8WXW0, Q8WXW1, Q8WXW2, Q96KJ1 | Gene names | WFDC2, HE4, WAP5 | |||
|
Domain Architecture |
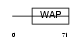
|
|||||
| Description | WAP four-disulfide core domain protein 2 precursor (Major epididymis- specific protein E4) (Epididymal secretory protein E4) (Putative protease inhibitor WAP5). | |||||
|
ALK1_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 23) | NC score | 0.512662 (rank : 17) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P03973, P07757 | Gene names | SLPI, WAP4, WFDC4 | |||
|
Domain Architecture |
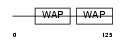
|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (HUSI-1) (Seminal proteinase inhibitor) (Secretory leukocyte protease inhibitor) (BLPI) (Mucus proteinase inhibitor) (MPI) (WAP four-disulfide core domain protein 4) (Protease inhibitor WAP4). | |||||
|
SPIT1_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 24) | NC score | 0.458979 (rank : 21) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
CO6A3_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 25) | NC score | 0.210589 (rank : 32) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
KALM_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 26) | NC score | 0.262402 (rank : 31) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P23352 | Gene names | KAL1, ADMLX, KAL, KALIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Anosmin-1 precursor (Kallmann syndrome protein) (Adhesion molecule- like X-linked). | |||||
|
SPIT3_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 27) | NC score | 0.656307 (rank : 5) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49223, Q6UDR8, Q96KK2 | Gene names | SPINT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kunitz-type protease inhibitor 3 precursor (HKIB9). | |||||
|
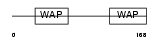
WFDC2_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 28) | NC score | 0.426045 (rank : 25) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9DAU7 | Gene names | Wfdc2, He4 | |||
|
Domain Architecture |
|
|||||
| Description | WAP four-disulfide core domain protein 2 precursor (WAP domain- containing protein HE4). | |||||
|
TFPI1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 29) | NC score | 0.555473 (rank : 12) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54819, Q9Z2U8 | Gene names | Tfpi | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
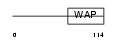
ELAF_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 30) | NC score | 0.458568 (rank : 22) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19957 | Gene names | PI3, WAP3, WFDC14 | |||
|
Domain Architecture |
|
|||||
| Description | Elafin precursor (Elastase-specific inhibitor) (ESI) (Skin-derived antileukoproteinase) (SKALP) (WAP four-disulfide core domain protein 14) (Protease inhibitor WAP3). | |||||
|
EXPI_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 31) | NC score | 0.434930 (rank : 24) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P62810, Q62477, Q91VQ6 | Gene names | Expi, Wdnm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular peptidase inhibitor precursor (Protein WDNM1). | |||||
|
WFD12_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 32) | NC score | 0.421040 (rank : 26) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JHY3 | Gene names | Wfdc12, Swam2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 12 precursor (Whey acidic protein 2) (Single WAP motif protein 2) (Elafin-like protein II). | |||||
|
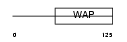
WAP_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 33) | NC score | 0.366282 (rank : 30) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P01173, P70230, Q61023 | Gene names | Wap | |||
|
Domain Architecture |
|
|||||
| Description | Whey acidic protein precursor (WAP). | |||||
|
NOTC1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 34) | NC score | 0.030857 (rank : 91) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
UROL1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 35) | NC score | 0.141347 (rank : 36) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5DID3, Q5DID1, Q5DID2 | Gene names | Umodl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
MCSP_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 36) | NC score | 0.156282 (rank : 33) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
KRA92_HUMAN
|
||||||
| θ value | 0.125558 (rank : 37) | NC score | 0.120861 (rank : 41) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.035312 (rank : 86) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
UROL1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.124701 (rank : 38) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5DID0, Q5DIC9, Q6LA40, Q6LA41, Q8N216 | Gene names | UMODL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
VWF_MOUSE
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.084000 (rank : 51) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
KRA98_HUMAN
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.109419 (rank : 44) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
ATS20_MOUSE
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.016611 (rank : 100) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59511 | Gene names | Adamts20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
ADA17_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.032800 (rank : 88) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P78536, O60226 | Gene names | ADAM17, CSVP, TACE | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (Snake venom-like protease) (CD156b antigen). | |||||
|
KR413_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.083066 (rank : 53) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYU7 | Gene names | KRTAP4-13, KAP4.13, KRTAP4.13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-13 (Keratin-associated protein 4.13) (Ultrahigh sulfur keratin-associated protein 4.13). | |||||
|
IGF1R_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.009105 (rank : 102) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P08069 | Gene names | IGF1R | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
KR106_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.080540 (rank : 55) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KRA45_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.082790 (rank : 54) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
LRP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.034251 (rank : 87) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
GRN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.084586 (rank : 49) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P28798 | Gene names | Grn | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7]. | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.060284 (rank : 76) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
SLIT3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.017128 (rank : 98) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WVB4 | Gene names | Slit3 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Slit3). | |||||
|
KRA44_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.084342 (rank : 50) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
KRA93_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.108090 (rank : 45) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
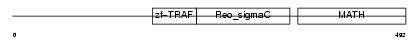
TRAF2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.017113 (rank : 99) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12933, Q96NT2 | Gene names | TRAF2, TRAP3 | |||
|
Domain Architecture |
|
|||||
| Description | TNF receptor-associated factor 2 (Tumor necrosis factor type 2 receptor-associated protein 3). | |||||
|
KR104_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.067212 (rank : 62) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KRA99_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.102591 (rank : 47) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
PCSK5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.032372 (rank : 89) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
WFDC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.138513 (rank : 37) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HC57, Q8NC27, Q9HAU1 | Gene names | WFDC1, PS20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 1 precursor (Prostate stromal protein ps20) (ps20 growth inhibitor). | |||||
|
WFDC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.142231 (rank : 35) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ESH5, Q8R110 | Gene names | Wfdc1, Ps20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 1 precursor (Prostate stromal protein ps20) (ps20 growth inhibitor). | |||||
|
ENPP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.030421 (rank : 92) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
KR10B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.075146 (rank : 59) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
ENPP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.029104 (rank : 93) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
GRN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.076066 (rank : 58) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
KR107_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.076300 (rank : 57) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KRA94_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.107905 (rank : 46) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BYQ2 | Gene names | KRTAP9-4, KAP9.4, KRTAP9.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-4 (Keratin-associated protein 9.4) (Ultrahigh sulfur keratin-associated protein 9.4). | |||||
|
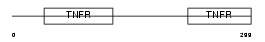
TNR8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.031658 (rank : 90) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
GLIP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.016158 (rank : 101) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CWG1 | Gene names | Glipr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma pathogenesis-related protein 1 precursor (GliPR 1). | |||||
|
KR10C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.064100 (rank : 69) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
MCSP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.083774 (rank : 52) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49901, Q96A42 | Gene names | SMCP, MCS, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
TMPSD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.002036 (rank : 106) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
WFD13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.119166 (rank : 42) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IUB5, Q5TEU7, Q8WWK7 | Gene names | WFDC13, C20orf138, WAP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein WFDC13 precursor. | |||||
|
BTAF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.007512 (rank : 104) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.064324 (rank : 68) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
RSPO2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.025062 (rank : 95) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BFU0, Q7TPX3 | Gene names | Rspo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-2 precursor (Roof plate-specific spondin-2) (Cysteine-rich and single thrombospondin domain-containing protein 2) (Cristin-2) (mCristin-2). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.047184 (rank : 85) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
VWF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.077691 (rank : 56) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
WF10A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.097496 (rank : 48) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H1F0, Q5TGZ7 | Gene names | WFDC10A, C20orf146, WAP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 10A precursor (Putative protease inhibitor WAP10A). | |||||
|
IGF1R_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.006328 (rank : 105) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60751, O70438, Q62123 | Gene names | Igf1r | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
NOTC4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.025797 (rank : 94) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.023179 (rank : 97) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RSPO2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.023625 (rank : 96) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6UXX9, Q4G0U4, Q8N6X6 | Gene names | RSPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-2 precursor (Roof plate-specific spondin-2) (hRspo2). | |||||
|
STAG3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.008486 (rank : 103) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70576 | Gene names | Stag3 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-3 (Stromal antigen 3) (Stromalin 3) (SCC3 homolog 3). | |||||
|
APLP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.123145 (rank : 39) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.122849 (rank : 40) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
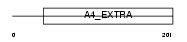
APLP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.146300 (rank : 34) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06335 | Gene names | Aplp2 | |||
|
Domain Architecture |
|
|||||
| Description | Amyloid-like protein 2 precursor (CDEI box-binding protein) (CDEBP). | |||||
|
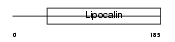
CO8G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.064954 (rank : 65) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07360, Q5SQ07 | Gene names | C8G | |||
|
Domain Architecture |
|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
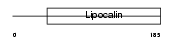
CO8G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.068607 (rank : 61) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCG4 | Gene names | C8g | |||
|
Domain Architecture |
|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
KR102_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.058638 (rank : 77) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR103_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.056623 (rank : 79) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
KR105_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.064930 (rank : 66) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KR108_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.050624 (rank : 84) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KR10A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.060685 (rank : 75) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60014 | Gene names | KRTAP10-10, KAP10.10, KAP18-10, KRTAP10.10, KRTAP18-1, KRTAP18.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-10 (Keratin-associated protein 10.10) (High sulfur keratin-associated protein 10.10) (Keratin-associated protein 18-10) (Keratin-associated protein 18.10). | |||||
|
KR111_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.053934 (rank : 82) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IUC1 | Gene names | KRTAP11-1, KAP11.1, KRTAP11.1 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin-associated protein 11-1 (High sulfur keratin-associated protein 11.1). | |||||
|
KR123_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.054172 (rank : 81) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60328 | Gene names | KRTAP12-3, KAP12.3, KRTAP12.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-3 (Keratin-associated protein 12.3) (High sulfur keratin-associated protein 12.3). | |||||
|
KR124_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.062282 (rank : 72) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P60329 | Gene names | KRTAP12-4, KAP12.4, KRTAP12.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-4 (Keratin-associated protein 12.4) (High sulfur keratin-associated protein 12.4). | |||||
|
KR410_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.062197 (rank : 73) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
KR412_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.073688 (rank : 60) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BQ66, Q495I0 | Gene names | KRTAP4-12, KAP4.12, KRTAP4.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-12 (Keratin-associated protein 4.12) (Ultrahigh sulfur keratin-associated protein 4.12). | |||||
|
KR414_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.065722 (rank : 63) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KRA24_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.065067 (rank : 64) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
KRA42_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.063527 (rank : 70) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYR5 | Gene names | KRTAP4-2, KAP4.2, KRTAP4.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-2 (Keratin-associated protein 4.2) (Ultrahigh sulfur keratin-associated protein 4.2). | |||||
|
KRA47_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.061028 (rank : 74) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.055845 (rank : 80) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
LRP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.063442 (rank : 71) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86VZ4, Q5VYC0, Q96SN6 | Gene names | LRP11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
LRP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.064407 (rank : 67) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CB67, Q8C7Y7 | Gene names | Lrp11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
PTGDS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.057086 (rank : 78) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41222, Q5SQ10, Q7M4P3, Q9UCC9, Q9UCD9 | Gene names | PTGDS, PDS | |||
|
Domain Architecture |
|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS) (Beta-trace protein) (Cerebrin-28). | |||||
|
PTGDS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.053819 (rank : 83) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09114, O09157, O35091, Q62169 | Gene names | Ptgds | |||
|
Domain Architecture |
|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS). | |||||
|
WFDC8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.05497e-141 (rank : 1) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
EPPI_MOUSE
|
||||||
| NC score | 0.827880 (rank : 2) | θ value | 9.56915e-18 (rank : 3) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9DA01 | Gene names | Spinlw1 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1). | |||||
|
EPPI_HUMAN
|
||||||
| NC score | 0.817635 (rank : 3) | θ value | 5.60996e-18 (rank : 2) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95925, Q86TP9, Q96SD7, Q9HD30 | Gene names | SPINLW1, WAP7, WFDC7 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1) (WAP four-disulfide core domain protein 7) (Protease inhibitor WAP7). | |||||
|
WFDC6_HUMAN
|
||||||
| NC score | 0.662989 (rank : 4) | θ value | 3.08544e-08 (rank : 7) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BQY6, Q5JYQ4, Q8NFV6 | Gene names | WFDC6, C20orf171, WAP6 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 6 precursor (Putative protease inhibitor WAP6). | |||||
|
SPIT3_HUMAN
|
||||||
| NC score | 0.656307 (rank : 5) | θ value | 0.00020696 (rank : 27) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49223, Q6UDR8, Q96KK2 | Gene names | SPINT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kunitz-type protease inhibitor 3 precursor (HKIB9). | |||||
|
TFPI2_MOUSE
|
||||||
| NC score | 0.613537 (rank : 6) | θ value | 4.0297e-08 (rank : 9) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35536 | Gene names | Tfpi2 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2). | |||||
|
TFPI2_HUMAN
|
||||||
| NC score | 0.612926 (rank : 7) | θ value | 4.0297e-08 (rank : 8) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P48307 | Gene names | TFPI2 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2) (Placental protein 5) (PP5). | |||||
|
SPIT2_MOUSE
|
||||||
| NC score | 0.592829 (rank : 8) | θ value | 4.92598e-06 (rank : 19) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WU03, Q9WU04, Q9WU05 | Gene names | Spint2, Hai2 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2). | |||||
|
SPIT2_HUMAN
|
||||||
| NC score | 0.592163 (rank : 9) | θ value | 1.69304e-06 (rank : 18) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43291, O00271, O14895, Q5TZQ3, Q969E0 | Gene names | SPINT2, HAI2, KOP | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2) (Placental bikunin). | |||||
|
WFD15_MOUSE
|
||||||
| NC score | 0.580176 (rank : 10) | θ value | 2.13673e-09 (rank : 6) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JHY4, Q8BVC0 | Gene names | Wfdc15, Swam1 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 15 precursor (Single WAP motif protein 1) (Elafin-like protein I). | |||||
|
TFPI1_HUMAN
|
||||||
| NC score | 0.579843 (rank : 11) | θ value | 1.29631e-06 (rank : 17) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P10646, O95103 | Gene names | TFPI, LACI, TFPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
TFPI1_MOUSE
|
||||||
| NC score | 0.555473 (rank : 12) | θ value | 0.000461057 (rank : 29) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54819, Q9Z2U8 | Gene names | Tfpi | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
WFDC5_HUMAN
|
||||||
| NC score | 0.543905 (rank : 13) | θ value | 7.34386e-10 (rank : 5) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TCV5, Q6UWE4 | Gene names | WFDC5, WAP1 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 5 precursor (Putative protease inhibitor WAP1). | |||||
|
ALK1_MOUSE
|
||||||
| NC score | 0.528174 (rank : 14) | θ value | 1.99992e-07 (rank : 12) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97430, O09081, O09082 | Gene names | Slpi | |||
|
Domain Architecture |

|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (Secretory leukocyte protease inhibitor). | |||||
|
WFD12_HUMAN
|
||||||
| NC score | 0.518496 (rank : 15) | θ value | 1.09739e-05 (rank : 21) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWY7, Q5H980, Q9BR31 | Gene names | WFDC12, C20orf122, WAP2 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 12 precursor (Putative protease inhibitor WAP12) (Whey acidic protein 2). | |||||
|
AMBP_HUMAN
|
||||||
| NC score | 0.516207 (rank : 16) | θ value | 8.97725e-08 (rank : 10) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P02760, P00977, P02759 | Gene names | AMBP, HCP, ITIL | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin (Protein HC) (Complex-forming glycoprotein heterogeneous in charge) (Alpha-1 microglycoprotein); Inter-alpha-trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
ALK1_HUMAN
|
||||||
| NC score | 0.512662 (rank : 17) | θ value | 1.43324e-05 (rank : 23) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P03973, P07757 | Gene names | SLPI, WAP4, WFDC4 | |||
|
Domain Architecture |

|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (HUSI-1) (Seminal proteinase inhibitor) (Secretory leukocyte protease inhibitor) (BLPI) (Mucus proteinase inhibitor) (MPI) (WAP four-disulfide core domain protein 4) (Protease inhibitor WAP4). | |||||
|
AMBP_MOUSE
|
||||||
| NC score | 0.499580 (rank : 18) | θ value | 9.92553e-07 (rank : 15) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
SPIT1_MOUSE
|
||||||
| NC score | 0.485813 (rank : 19) | θ value | 8.97725e-08 (rank : 11) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9R097 | Gene names | Spint1, Hai1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
WFDC3_HUMAN
|
||||||
| NC score | 0.466777 (rank : 20) | θ value | 6.64225e-11 (rank : 4) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IUB2, Q8TC52, Q9BQP3, Q9BQP4 | Gene names | WFDC3, WAP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 3 precursor (Putative protease inhibitor WAP14). | |||||
|
SPIT1_HUMAN
|
||||||
| NC score | 0.458979 (rank : 21) | θ value | 1.43324e-05 (rank : 24) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
ELAF_HUMAN
|
||||||
| NC score | 0.458568 (rank : 22) | θ value | 0.000786445 (rank : 30) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19957 | Gene names | PI3, WAP3, WFDC14 | |||
|
Domain Architecture |

|
|||||
| Description | Elafin precursor (Elastase-specific inhibitor) (ESI) (Skin-derived antileukoproteinase) (SKALP) (WAP four-disulfide core domain protein 14) (Protease inhibitor WAP3). | |||||
|
WFDC2_HUMAN
|
||||||
| NC score | 0.448302 (rank : 23) | θ value | 1.09739e-05 (rank : 22) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14508, Q8WXV9, Q8WXW0, Q8WXW1, Q8WXW2, Q96KJ1 | Gene names | WFDC2, HE4, WAP5 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 2 precursor (Major epididymis- specific protein E4) (Epididymal secretory protein E4) (Putative protease inhibitor WAP5). | |||||
|
EXPI_MOUSE
|
||||||
| NC score | 0.434930 (rank : 24) | θ value | 0.00390308 (rank : 31) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P62810, Q62477, Q91VQ6 | Gene names | Expi, Wdnm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular peptidase inhibitor precursor (Protein WDNM1). | |||||
|
WFDC2_MOUSE
|
||||||
| NC score | 0.426045 (rank : 25) | θ value | 0.00035302 (rank : 28) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9DAU7 | Gene names | Wfdc2, He4 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 2 precursor (WAP domain- containing protein HE4). | |||||
|
WFD12_MOUSE
|
||||||
| NC score | 0.421040 (rank : 26) | θ value | 0.00390308 (rank : 32) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JHY3 | Gene names | Wfdc12, Swam2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 12 precursor (Whey acidic protein 2) (Single WAP motif protein 2) (Elafin-like protein II). | |||||
|
A4_HUMAN
|
||||||
| NC score | 0.419639 (rank : 27) | θ value | 7.59969e-07 (rank : 13) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.419530 (rank : 28) | θ value | 7.59969e-07 (rank : 14) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
APLP2_HUMAN
|
||||||
| NC score | 0.416018 (rank : 29) | θ value | 9.92553e-07 (rank : 16) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
WAP_MOUSE
|
||||||
| NC score | 0.366282 (rank : 30) | θ value | 0.0113563 (rank : 33) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P01173, P70230, Q61023 | Gene names | Wap | |||
|
Domain Architecture |

|
|||||
| Description | Whey acidic protein precursor (WAP). | |||||
|
KALM_HUMAN
|
||||||
| NC score | 0.262402 (rank : 31) | θ value | 0.000121331 (rank : 26) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P23352 | Gene names | KAL1, ADMLX, KAL, KALIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Anosmin-1 precursor (Kallmann syndrome protein) (Adhesion molecule- like X-linked). | |||||
|
CO6A3_HUMAN
|
||||||
| NC score | 0.210589 (rank : 32) | θ value | 9.29e-05 (rank : 25) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
MCSP_MOUSE
|
||||||
| NC score | 0.156282 (rank : 33) | θ value | 0.0961366 (rank : 36) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
APLP2_MOUSE
|
||||||
| NC score | 0.146300 (rank : 34) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06335 | Gene names | Aplp2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (CDEI box-binding protein) (CDEBP). | |||||
|
WFDC1_MOUSE
|
||||||
| NC score | 0.142231 (rank : 35) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ESH5, Q8R110 | Gene names | Wfdc1, Ps20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 1 precursor (Prostate stromal protein ps20) (ps20 growth inhibitor). | |||||
|
UROL1_MOUSE
|
||||||
| NC score | 0.141347 (rank : 36) | θ value | 0.0330416 (rank : 35) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5DID3, Q5DID1, Q5DID2 | Gene names | Umodl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
WFDC1_HUMAN
|
||||||
| NC score | 0.138513 (rank : 37) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HC57, Q8NC27, Q9HAU1 | Gene names | WFDC1, PS20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 1 precursor (Prostate stromal protein ps20) (ps20 growth inhibitor). | |||||
|
UROL1_HUMAN
|
||||||
| NC score | 0.124701 (rank : 38) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5DID0, Q5DIC9, Q6LA40, Q6LA41, Q8N216 | Gene names | UMODL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
APLP1_HUMAN
|
||||||
| NC score | 0.123145 (rank : 39) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP1_MOUSE
|
||||||
| NC score | 0.122849 (rank : 40) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
KRA92_HUMAN
|
||||||
| NC score | 0.120861 (rank : 41) | θ value | 0.125558 (rank : 37) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
WFD13_HUMAN
|
||||||
| NC score | 0.119166 (rank : 42) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IUB5, Q5TEU7, Q8WWK7 | Gene names | WFDC13, C20orf138, WAP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein WFDC13 precursor. | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.118275 (rank : 43) | θ value | 1.09739e-05 (rank : 20) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
KRA98_HUMAN
|
||||||
| NC score | 0.109419 (rank : 44) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
KRA93_HUMAN
|
||||||
| NC score | 0.108090 (rank : 45) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
KRA94_HUMAN
|
||||||
| NC score | 0.107905 (rank : 46) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BYQ2 | Gene names | KRTAP9-4, KAP9.4, KRTAP9.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-4 (Keratin-associated protein 9.4) (Ultrahigh sulfur keratin-associated protein 9.4). | |||||
|
KRA99_HUMAN
|
||||||
| NC score | 0.102591 (rank : 47) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
WF10A_HUMAN
|
||||||
| NC score | 0.097496 (rank : 48) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H1F0, Q5TGZ7 | Gene names | WFDC10A, C20orf146, WAP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 10A precursor (Putative protease inhibitor WAP10A). | |||||
|
GRN_MOUSE
|
||||||
| NC score | 0.084586 (rank : 49) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P28798 | Gene names | Grn | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7]. | |||||
|
KRA44_HUMAN
|
||||||
| NC score | 0.084342 (rank : 50) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.084000 (rank : 51) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
MCSP_HUMAN
|
||||||
| NC score | 0.083774 (rank : 52) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49901, Q96A42 | Gene names | SMCP, MCS, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
KR413_HUMAN
|
||||||
| NC score | 0.083066 (rank : 53) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYU7 | Gene names | KRTAP4-13, KAP4.13, KRTAP4.13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-13 (Keratin-associated protein 4.13) (Ultrahigh sulfur keratin-associated protein 4.13). | |||||
|
KRA45_HUMAN
|
||||||
| NC score | 0.082790 (rank : 54) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
KR106_HUMAN
|
||||||
| NC score | 0.080540 (rank : 55) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
VWF_HUMAN
|
||||||
| NC score | 0.077691 (rank : 56) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.076300 (rank : 57) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
GRN_HUMAN
|
||||||
| NC score | 0.076066 (rank : 58) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
KR10B_HUMAN
|
||||||
| NC score | 0.075146 (rank : 59) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
KR412_HUMAN
|
||||||
| NC score | 0.073688 (rank : 60) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BQ66, Q495I0 | Gene names | KRTAP4-12, KAP4.12, KRTAP4.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-12 (Keratin-associated protein 4.12) (Ultrahigh sulfur keratin-associated protein 4.12). | |||||
|
CO8G_MOUSE
|
||||||
| NC score | 0.068607 (rank : 61) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCG4 | Gene names | C8g | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.067212 (rank : 62) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KR414_HUMAN
|
||||||
| NC score | 0.065722 (rank : 63) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KRA24_HUMAN
|
||||||
| NC score | 0.065067 (rank : 64) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
CO8G_HUMAN
|
||||||
| NC score | 0.064954 (rank : 65) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07360, Q5SQ07 | Gene names | C8G | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.064930 (rank : 66) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
LRP11_MOUSE
|
||||||
| NC score | 0.064407 (rank : 67) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CB67, Q8C7Y7 | Gene names | Lrp11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.064324 (rank : 68) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KR10C_HUMAN
|
||||||
| NC score | 0.064100 (rank : 69) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KRA42_HUMAN
|
||||||
| NC score | 0.063527 (rank : 70) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYR5 | Gene names | KRTAP4-2, KAP4.2, KRTAP4.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-2 (Keratin-associated protein 4.2) (Ultrahigh sulfur keratin-associated protein 4.2). | |||||
|
LRP11_HUMAN
|
||||||
| NC score | 0.063442 (rank : 71) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86VZ4, Q5VYC0, Q96SN6 | Gene names | LRP11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
KR124_HUMAN
|
||||||
| NC score | 0.062282 (rank : 72) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P60329 | Gene names | KRTAP12-4, KAP12.4, KRTAP12.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-4 (Keratin-associated protein 12.4) (High sulfur keratin-associated protein 12.4). | |||||
|
KR410_HUMAN
|
||||||
| NC score | 0.062197 (rank : 73) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
KRA47_HUMAN
|
||||||
| NC score | 0.061028 (rank : 74) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
KR10A_HUMAN
|
||||||
| NC score | 0.060685 (rank : 75) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60014 | Gene names | KRTAP10-10, KAP10.10, KAP18-10, KRTAP10.10, KRTAP18-1, KRTAP18.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-10 (Keratin-associated protein 10.10) (High sulfur keratin-associated protein 10.10) (Keratin-associated protein 18-10) (Keratin-associated protein 18.10). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.060284 (rank : 76) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
KR102_HUMAN
|
||||||
| NC score | 0.058638 (rank : 77) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
PTGDS_HUMAN
|
||||||
| NC score | 0.057086 (rank : 78) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41222, Q5SQ10, Q7M4P3, Q9UCC9, Q9UCD9 | Gene names | PTGDS, PDS | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS) (Beta-trace protein) (Cerebrin-28). | |||||
|
KR103_HUMAN
|
||||||
| NC score | 0.056623 (rank : 79) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.055845 (rank : 80) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KR123_HUMAN
|
||||||
| NC score | 0.054172 (rank : 81) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60328 | Gene names | KRTAP12-3, KAP12.3, KRTAP12.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-3 (Keratin-associated protein 12.3) (High sulfur keratin-associated protein 12.3). | |||||
|
KR111_HUMAN
|
||||||
| NC score | 0.053934 (rank : 82) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IUC1 | Gene names | KRTAP11-1, KAP11.1, KRTAP11.1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin-associated protein 11-1 (High sulfur keratin-associated protein 11.1). | |||||
|
PTGDS_MOUSE
|
||||||
| NC score | 0.053819 (rank : 83) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09114, O09157, O35091, Q62169 | Gene names | Ptgds | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS). | |||||
|
KR108_HUMAN
|
||||||
| NC score | 0.050624 (rank : 84) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.047184 (rank : 85) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.035312 (rank : 86) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.034251 (rank : 87) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
ADA17_HUMAN
|
||||||
| NC score | 0.032800 (rank : 88) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P78536, O60226 | Gene names | ADAM17, CSVP, TACE | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (Snake venom-like protease) (CD156b antigen). | |||||
|
PCSK5_MOUSE
|
||||||
| NC score | 0.032372 (rank : 89) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
TNR8_HUMAN
|
||||||
| NC score | 0.031658 (rank : 90) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
NOTC1_HUMAN
|
||||||
| NC score | 0.030857 (rank : 91) | θ value | 0.0330416 (rank : 34) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
ENPP2_HUMAN
|
||||||
| NC score | 0.030421 (rank : 92) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_MOUSE
|
||||||
| NC score | 0.029104 (rank : 93) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
NOTC4_MOUSE
|
||||||
| NC score | 0.025797 (rank : 94) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
RSPO2_MOUSE
|
||||||
| NC score | 0.025062 (rank : 95) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BFU0, Q7TPX3 | Gene names | Rspo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-2 precursor (Roof plate-specific spondin-2) (Cysteine-rich and single thrombospondin domain-containing protein 2) (Cristin-2) (mCristin-2). | |||||
|
RSPO2_HUMAN
|
||||||
| NC score | 0.023625 (rank : 96) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6UXX9, Q4G0U4, Q8N6X6 | Gene names | RSPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-2 precursor (Roof plate-specific spondin-2) (hRspo2). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.023179 (rank : 97) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SLIT3_MOUSE
|
||||||
| NC score | 0.017128 (rank : 98) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WVB4 | Gene names | Slit3 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Slit3). | |||||
|
TRAF2_HUMAN
|
||||||
| NC score | 0.017113 (rank : 99) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12933, Q96NT2 | Gene names | TRAF2, TRAP3 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 2 (Tumor necrosis factor type 2 receptor-associated protein 3). | |||||
|
ATS20_MOUSE
|
||||||
| NC score | 0.016611 (rank : 100) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59511 | Gene names | Adamts20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
GLIP1_MOUSE
|
||||||
| NC score | 0.016158 (rank : 101) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CWG1 | Gene names | Glipr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma pathogenesis-related protein 1 precursor (GliPR 1). | |||||
|
IGF1R_HUMAN
|
||||||
| NC score | 0.009105 (rank : 102) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P08069 | Gene names | IGF1R | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
STAG3_MOUSE
|
||||||
| NC score | 0.008486 (rank : 103) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70576 | Gene names | Stag3 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-3 (Stromal antigen 3) (Stromalin 3) (SCC3 homolog 3). | |||||
|
BTAF1_HUMAN
|
||||||
| NC score | 0.007512 (rank : 104) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
IGF1R_MOUSE
|
||||||
| NC score | 0.006328 (rank : 105) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60751, O70438, Q62123 | Gene names | Igf1r | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
TMPSD_HUMAN
|
||||||
| NC score | 0.002036 (rank : 106) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||