Please be patient as the page loads
|
MCSP_HUMAN
|
||||||
| SwissProt Accessions | P49901, Q96A42 | Gene names | SMCP, MCS, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MCSP_HUMAN
|
||||||
| θ value | 1.79215e-40 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P49901, Q96A42 | Gene names | SMCP, MCS, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 2) | NC score | 0.238105 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 3) | NC score | 0.247237 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
KR124_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 4) | NC score | 0.179161 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60329 | Gene names | KRTAP12-4, KAP12.4, KRTAP12.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-4 (Keratin-associated protein 12.4) (High sulfur keratin-associated protein 12.4). | |||||
|
GRN_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 5) | NC score | 0.269175 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P28798 | Gene names | Grn | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7]. | |||||
|
GRN_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 6) | NC score | 0.265726 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
MCSP_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 7) | NC score | 0.490398 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
ADA32_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.057847 (rank : 126) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K410, Q6P901, Q8BJ80 | Gene names | Adam32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAM 32 precursor (A disintegrin and metalloproteinase domain 32). | |||||
|
DLL3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.068315 (rank : 112) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
KR412_HUMAN
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.148328 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BQ66, Q495I0 | Gene names | KRTAP4-12, KAP4.12, KRTAP4.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-12 (Keratin-associated protein 4.12) (Ultrahigh sulfur keratin-associated protein 4.12). | |||||
|
ERBB3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.025720 (rank : 161) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P21860 | Gene names | ERBB3, HER3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Tyrosine kinase-type cell surface receptor HER3). | |||||
|
SPR1A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 12) | NC score | 0.235342 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.077433 (rank : 101) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
LAMA5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.069812 (rank : 109) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LSR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.111736 (rank : 66) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86X29, O00112, O00426, Q6ZT80, Q9UQL3 | Gene names | LSR, LISCH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipolysis-stimulated lipoprotein receptor. | |||||
|
LSR_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.114832 (rank : 60) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99KG5, Q3TJE7, Q3UIQ9, Q61148, Q61149, Q6U816 | Gene names | Lsr, Lisch7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipolysis-stimulated lipoprotein receptor precursor (Liver-specific bHLH-Zip transcription factor) (Lipolysis-stimulated receptor) (Liver- specific gene on mouse chromosome 7 protein). | |||||
|
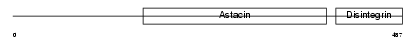
ADA10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.090069 (rank : 91) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14672, Q10742, Q92650 | Gene names | ADAM10, KUZ, MADM | |||
|
Domain Architecture |
|
|||||
| Description | ADAM 10 precursor (EC 3.4.24.81) (A disintegrin and metalloproteinase domain 10) (Mammalian disintegrin-metalloprotease) (Kuzbanian protein homolog) (CDw156c antigen). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.120271 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
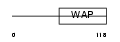
WFDC5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.137393 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TCV5, Q6UWE4 | Gene names | WFDC5, WAP1 | |||
|
Domain Architecture |
|
|||||
| Description | WAP four-disulfide core domain protein 5 precursor (Putative protease inhibitor WAP1). | |||||
|
ADA10_MOUSE
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.090107 (rank : 90) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O35598 | Gene names | Adam10, Kuz, Madm | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 10 precursor (EC 3.4.24.81) (A disintegrin and metalloproteinase domain 10) (Mammalian disintegrin-metalloprotease) (Kuzbanian protein homolog). | |||||
|
LRP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.060345 (rank : 122) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.081280 (rank : 99) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
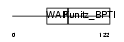
EPPI_MOUSE
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.060896 (rank : 121) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DA01 | Gene names | Spinlw1 | |||
|
Domain Architecture |
|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1). | |||||
|
FOLR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.052003 (rank : 148) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14207 | Gene names | FOLR2 | |||
|
Domain Architecture |

|
|||||
| Description | Folate receptor beta precursor (FR-beta) (Folate receptor 2) (Folate receptor, fetal/placental) (Placental folate-binding protein) (FBP). | |||||
|
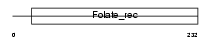
FOLR3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.052025 (rank : 147) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41439 | Gene names | FOLR3 | |||
|
Domain Architecture |
|
|||||
| Description | Folate receptor gamma precursor (FR-gamma) (Folate receptor 3). | |||||
|
KR105_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.139736 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KRA56_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.119087 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
NOTC4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.056738 (rank : 129) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
SPR1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.231461 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
ERBB3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.023378 (rank : 163) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 898 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61526, Q3KQR1, Q68J64, Q810U8, Q8K317 | Gene names | Erbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Glial growth factor receptor). | |||||
|
KR511_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.125769 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA52_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.109252 (rank : 70) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.116133 (rank : 56) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.039189 (rank : 160) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
WFDC8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.083774 (rank : 95) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |
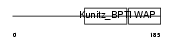
|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
ADA17_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.070612 (rank : 107) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P78536, O60226 | Gene names | ADAM17, CSVP, TACE | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (Snake venom-like protease) (CD156b antigen). | |||||
|
ALK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.071288 (rank : 106) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97430, O09081, O09082 | Gene names | Slpi | |||
|
Domain Architecture |
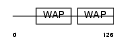
|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (Secretory leukocyte protease inhibitor). | |||||
|
FBN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.053219 (rank : 141) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
KR510_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.120740 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
KRA57_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.110498 (rank : 67) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
SREC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.089183 (rank : 93) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
TINAL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.024537 (rank : 162) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99JR5, Q9EQT9 | Gene names | Tinagl1, Arg1, Lcn7, Tinagl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Androgen- regulated gene 1 protein) (Adrenocortical zonation factor 1) (AZ-1) (Tubulointerstitial nephritis antigen-related protein) (TARP). | |||||
|
FBN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.052578 (rank : 145) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
KRA51_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.097147 (rank : 82) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.112051 (rank : 64) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
LCE3B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.117258 (rank : 52) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5TA77 | Gene names | LCE3B, LEP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3B (Late envelope protein 14). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.040008 (rank : 159) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
TNR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.022828 (rank : 164) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P36941 | Gene names | LTBR, TNFCR, TNFRSF3 | |||
|
Domain Architecture |
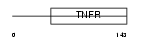
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor) (Tumor necrosis factor receptor 2-related protein) (Tumor necrosis factor C receptor). | |||||
|
ADA17_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.061260 (rank : 118) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z0F8, O88726, Q9R1U4, Q9Z0K3 | Gene names | Adam17, Tace | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (CD156b antigen). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.065646 (rank : 114) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
ALK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.058191 (rank : 125) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03973, P07757 | Gene names | SLPI, WAP4, WFDC4 | |||
|
Domain Architecture |
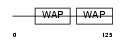
|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (HUSI-1) (Seminal proteinase inhibitor) (Secretory leukocyte protease inhibitor) (BLPI) (Mucus proteinase inhibitor) (MPI) (WAP four-disulfide core domain protein 4) (Protease inhibitor WAP4). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.056508 (rank : 131) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
DLK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.053013 (rank : 143) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.050312 (rank : 155) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
DLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.053551 (rank : 140) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
DLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.055995 (rank : 133) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
DLL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.050773 (rank : 153) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
FBN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.050247 (rank : 156) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.050606 (rank : 154) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.053831 (rank : 138) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
FRAS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.054927 (rank : 134) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
KR101_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.115450 (rank : 57) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR102_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.116497 (rank : 54) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR103_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.114627 (rank : 61) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
KR104_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.111829 (rank : 65) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KR106_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.118699 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR107_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.129735 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR108_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.107504 (rank : 73) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KR109_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.104964 (rank : 75) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
KR10A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.091666 (rank : 89) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P60014 | Gene names | KRTAP10-10, KAP10.10, KAP18-10, KRTAP10.10, KRTAP18-1, KRTAP18.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-10 (Keratin-associated protein 10.10) (High sulfur keratin-associated protein 10.10) (Keratin-associated protein 18-10) (Keratin-associated protein 18.10). | |||||
|
KR10B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.124265 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
KR10C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.094723 (rank : 84) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KR111_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.061207 (rank : 119) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IUC1 | Gene names | KRTAP11-1, KAP11.1, KRTAP11.1 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin-associated protein 11-1 (High sulfur keratin-associated protein 11.1). | |||||
|
KR121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.085856 (rank : 94) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59990 | Gene names | KRTAP12-1, KAP12.1, KRTAP12.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-1 (Keratin-associated protein 12.1) (High sulfur keratin-associated protein 12.1). | |||||
|
KR122_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.094206 (rank : 85) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P59991 | Gene names | KRTAP12-2, KAP12.2, KRTAP12.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-2 (Keratin-associated protein 12.2) (High sulfur keratin-associated protein 12.2). | |||||
|
KR123_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.092051 (rank : 88) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P60328 | Gene names | KRTAP12-3, KAP12.3, KRTAP12.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-3 (Keratin-associated protein 12.3) (High sulfur keratin-associated protein 12.3). | |||||
|
KR171_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.093480 (rank : 86) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
KR410_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.114568 (rank : 62) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
KR413_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.094914 (rank : 83) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYU7 | Gene names | KRTAP4-13, KAP4.13, KRTAP4.13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-13 (Keratin-associated protein 4.13) (Ultrahigh sulfur keratin-associated protein 4.13). | |||||
|
KR414_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.116876 (rank : 53) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KRA11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.104189 (rank : 76) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q07627, Q96S60, Q96S67 | Gene names | KRTAP1-1, B2A, KAP1.1, KAP1.6, KAP1.7, KRTAP1.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-1 (Keratin-associated protein 1.1) (High sulfur keratin-associated protein 1.1) (Keratin-associated protein 1.6) (Keratin-associated protein 1.7). | |||||
|
KRA13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.103371 (rank : 78) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IUG1, Q07628, Q8IUG0, Q9BYS2 | Gene names | KRTAP1-3, B2B, KAP1.2, KAP1.3, KAP1.8, KAP1.9, KRATP1.9, KRTAP1.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-3 (Keratin-associated protein 1.8) (Keratin-associated protein 1.9). | |||||
|
KRA15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.115342 (rank : 58) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYS1, Q52LP6 | Gene names | KRTAP1-5, KAP1.5, KRTAP1.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-5 (Keratin-associated protein 1.5) (High sulfur keratin-associated protein 1.5). | |||||
|
KRA24_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.116264 (rank : 55) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
KRA42_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.115012 (rank : 59) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BYR5 | Gene names | KRTAP4-2, KAP4.2, KRTAP4.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-2 (Keratin-associated protein 4.2) (Ultrahigh sulfur keratin-associated protein 4.2). | |||||
|
KRA44_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.107919 (rank : 72) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
KRA45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.121316 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
KRA47_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.114186 (rank : 63) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
KRA54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.098142 (rank : 81) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.083599 (rank : 96) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KRA92_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.105622 (rank : 74) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
KRA93_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.098454 (rank : 80) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
KRA94_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.108740 (rank : 71) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYQ2 | Gene names | KRTAP9-4, KAP9.4, KRTAP9.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-4 (Keratin-associated protein 9.4) (Ultrahigh sulfur keratin-associated protein 9.4). | |||||
|
KRA98_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.109283 (rank : 69) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
KRA99_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.089997 (rank : 92) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
LAMA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.063185 (rank : 115) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LCE1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.171810 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
LCE1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.174192 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
LCE1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.196693 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
LCE1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.151193 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
LCE1E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.209220 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LCE1F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.219398 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
LCE2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.277124 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
LCE2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.268512 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
LCE2C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.260493 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
LCE2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.263005 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
LCE3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.100211 (rank : 79) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5TA76 | Gene names | LCE3A, LEP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3A (Late envelope protein 13). | |||||
|
LCE3D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.127569 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYE3 | Gene names | LCE3D, LEP16, SPRL6A, SPRL6B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3D (Late envelope protein 16) (Small proline-rich-like epidermal differentiation complex protein 6A) (Small proline-rich-like epidermal differentiation complex protein 6B). | |||||
|
LCE3E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.092234 (rank : 87) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5T5B0 | Gene names | LCE3E, LEP17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3E (Late envelope protein 17). | |||||
|
LCE4A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.123701 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
LCE5A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.292372 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
LRP1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.053041 (rank : 142) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.051458 (rank : 150) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.051289 (rank : 151) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.068615 (rank : 111) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.053658 (rank : 139) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.075116 (rank : 104) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P25713, Q2V574 | Gene names | MT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-3 (MT-3) (Metallothionein-III) (MT-III) (Growth inhibitory factor) (GIF) (GIFB). | |||||
|
NAGPA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.056714 (rank : 130) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BJ48, Q3UUT5, Q8CHQ8, Q9QZE6 | Gene names | Nagpa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
NFX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.077457 (rank : 100) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
NICE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.053920 (rank : 137) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9UGL9 | Gene names | NICE1, C1orf42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein NICE-1. | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.061901 (rank : 116) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
ODFP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.068973 (rank : 110) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61999 | Gene names | Odf1, Odfp | |||
|
Domain Architecture |
|
|||||
| Description | Outer dense fiber protein. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.052153 (rank : 146) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.070377 (rank : 108) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.058421 (rank : 123) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.051459 (rank : 149) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.076031 (rank : 103) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRR13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.083141 (rank : 97) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PS1C2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.061304 (rank : 117) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.076789 (rank : 102) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
SPR2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.235277 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35326 | Gene names | SPRR2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A (SPR-2A) (2-1). | |||||
|
SPR2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.103409 (rank : 77) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CQK8, O70553 | Gene names | Sprr2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A. | |||||
|
SPR2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.258155 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35325, Q5T528 | Gene names | SPRR2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B (SPR-2B). | |||||
|
SPR2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.263158 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
SPR2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.263139 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22532, Q5T523, Q96RM3 | Gene names | SPRR2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D (SPR-2D) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.253365 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
SPR2E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.235527 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22531, Q96RM2 | Gene names | SPRR2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E (SPR-2E) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR2E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.239120 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
SPR2F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.223366 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96RM1 | Gene names | SPRR2F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F (SPR-2F). | |||||
|
SPR2F_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.233585 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
SPR2G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.256827 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYE4 | Gene names | SPRR2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G (SPR-2G). | |||||
|
SPR2G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.263984 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70558 | Gene names | Sprr2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G. | |||||
|
SPR2H_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.247312 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
SPR2I_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.226162 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
SPR2K_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.226115 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70562 | Gene names | Sprr2k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2K. | |||||
|
SPRR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.159601 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
SPRR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.149741 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
SPRR4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.140708 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96PI1, Q2M1Y7, Q5T522 | Gene names | SPRR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
SPRR4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.118071 (rank : 51) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CGN8 | Gene names | Sprr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
SREC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.081538 (rank : 98) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.056826 (rank : 128) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
STAB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.054281 (rank : 136) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
STAB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.054421 (rank : 135) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
STAB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.050062 (rank : 158) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
TENX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.050801 (rank : 152) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
TROAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.065759 (rank : 113) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
VWF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.050243 (rank : 157) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
WFDC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.060914 (rank : 120) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14508, Q8WXV9, Q8WXW0, Q8WXW1, Q8WXW2, Q96KJ1 | Gene names | WFDC2, HE4, WAP5 | |||
|
Domain Architecture |
|
|||||
| Description | WAP four-disulfide core domain protein 2 precursor (Major epididymis- specific protein E4) (Epididymal secretory protein E4) (Putative protease inhibitor WAP5). | |||||
|
WFDC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.110348 (rank : 68) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IUB2, Q8TC52, Q9BQP3, Q9BQP4 | Gene names | WFDC3, WAP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 3 precursor (Putative protease inhibitor WAP14). | |||||
|
WIF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.056866 (rank : 127) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
WIF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.058357 (rank : 124) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.071806 (rank : 105) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZNFX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.052680 (rank : 144) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
ZNFX1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.056075 (rank : 132) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
MCSP_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.79215e-40 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P49901, Q96A42 | Gene names | SMCP, MCS, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
MCSP_MOUSE
|
||||||
| NC score | 0.490398 (rank : 2) | θ value | 0.00509761 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
LCE5A_HUMAN
|
||||||
| NC score | 0.292372 (rank : 3) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
LCE2A_HUMAN
|
||||||
| NC score | 0.277124 (rank : 4) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
GRN_MOUSE
|
||||||
| NC score | 0.269175 (rank : 5) | θ value | 0.00228821 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P28798 | Gene names | Grn | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7]. | |||||
|
LCE2B_HUMAN
|
||||||
| NC score | 0.268512 (rank : 6) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
GRN_HUMAN
|
||||||
| NC score | 0.265726 (rank : 7) | θ value | 0.00298849 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
SPR2G_MOUSE
|
||||||
| NC score | 0.263984 (rank : 8) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70558 | Gene names | Sprr2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G. | |||||
|
SPR2B_MOUSE
|
||||||
| NC score | 0.263158 (rank : 9) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
SPR2D_HUMAN
|
||||||
| NC score | 0.263139 (rank : 10) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22532, Q5T523, Q96RM3 | Gene names | SPRR2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D (SPR-2D) (Small proline-rich protein II) (SPR-II). | |||||
|
LCE2D_HUMAN
|
||||||
| NC score | 0.263005 (rank : 11) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
LCE2C_HUMAN
|
||||||
| NC score | 0.260493 (rank : 12) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
SPR2B_HUMAN
|
||||||
| NC score | 0.258155 (rank : 13) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35325, Q5T528 | Gene names | SPRR2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B (SPR-2B). | |||||
|
SPR2G_HUMAN
|
||||||
| NC score | 0.256827 (rank : 14) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYE4 | Gene names | SPRR2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G (SPR-2G). | |||||
|
SPR2D_MOUSE
|
||||||
| NC score | 0.253365 (rank : 15) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
SPR2H_MOUSE
|
||||||
| NC score | 0.247312 (rank : 16) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70559 | Gene names | Sprr2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2H. | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.247237 (rank : 17) | θ value | 9.29e-05 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SPR2E_MOUSE
|
||||||
| NC score | 0.239120 (rank : 18) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.238105 (rank : 19) | θ value | 7.1131e-05 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SPR2E_HUMAN
|
||||||
| NC score | 0.235527 (rank : 20) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22531, Q96RM2 | Gene names | SPRR2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E (SPR-2E) (Small proline-rich protein II) (SPR-II). | |||||
|
SPR1A_HUMAN
|
||||||
| NC score | 0.235342 (rank : 21) | θ value | 0.813845 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
SPR2A_HUMAN
|
||||||
| NC score | 0.235277 (rank : 22) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35326 | Gene names | SPRR2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A (SPR-2A) (2-1). | |||||
|
SPR2F_MOUSE
|
||||||
| NC score | 0.233585 (rank : 23) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
SPR1B_HUMAN
|
||||||
| NC score | 0.231461 (rank : 24) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
SPR2I_MOUSE
|
||||||
| NC score | 0.226162 (rank : 25) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
SPR2K_MOUSE
|
||||||
| NC score | 0.226115 (rank : 26) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70562 | Gene names | Sprr2k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2K. | |||||
|
SPR2F_HUMAN
|
||||||
| NC score | 0.223366 (rank : 27) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96RM1 | Gene names | SPRR2F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F (SPR-2F). | |||||
|
LCE1F_HUMAN
|
||||||
| NC score | 0.219398 (rank : 28) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
LCE1E_HUMAN
|
||||||
| NC score | 0.209220 (rank : 29) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LCE1C_HUMAN
|
||||||
| NC score | 0.196693 (rank : 30) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
KR124_HUMAN
|
||||||
| NC score | 0.179161 (rank : 31) | θ value | 0.00175202 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60329 | Gene names | KRTAP12-4, KAP12.4, KRTAP12.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-4 (Keratin-associated protein 12.4) (High sulfur keratin-associated protein 12.4). | |||||
|
LCE1B_HUMAN
|
||||||
| NC score | 0.174192 (rank : 32) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
LCE1A_HUMAN
|
||||||
| NC score | 0.171810 (rank : 33) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
SPRR3_HUMAN
|
||||||
| NC score | 0.159601 (rank : 34) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
LCE1D_HUMAN
|
||||||
| NC score | 0.151193 (rank : 35) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
SPRR3_MOUSE
|
||||||
| NC score | 0.149741 (rank : 36) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
KR412_HUMAN
|
||||||
| NC score | 0.148328 (rank : 37) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BQ66, Q495I0 | Gene names | KRTAP4-12, KAP4.12, KRTAP4.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-12 (Keratin-associated protein 4.12) (Ultrahigh sulfur keratin-associated protein 4.12). | |||||
|
SPRR4_HUMAN
|
||||||
| NC score | 0.140708 (rank : 38) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96PI1, Q2M1Y7, Q5T522 | Gene names | SPRR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.139736 (rank : 39) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
WFDC5_HUMAN
|
||||||
| NC score | 0.137393 (rank : 40) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TCV5, Q6UWE4 | Gene names | WFDC5, WAP1 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 5 precursor (Putative protease inhibitor WAP1). | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.129735 (rank : 41) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
LCE3D_HUMAN
|
||||||
| NC score | 0.127569 (rank : 42) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYE3 | Gene names | LCE3D, LEP16, SPRL6A, SPRL6B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3D (Late envelope protein 16) (Small proline-rich-like epidermal differentiation complex protein 6A) (Small proline-rich-like epidermal differentiation complex protein 6B). | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.125769 (rank : 43) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KR10B_HUMAN
|
||||||
| NC score | 0.124265 (rank : 44) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
LCE4A_HUMAN
|
||||||
| NC score | 0.123701 (rank : 45) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
KRA45_HUMAN
|
||||||
| NC score | 0.121316 (rank : 46) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
KR510_HUMAN
|
||||||
| NC score | 0.120740 (rank : 47) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.120271 (rank : 48) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KRA56_HUMAN
|
||||||
| NC score | 0.119087 (rank : 49) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KR106_HUMAN
|
||||||
| NC score | 0.118699 (rank : 50) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
SPRR4_MOUSE
|
||||||
| NC score | 0.118071 (rank : 51) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CGN8 | Gene names | Sprr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 4. | |||||
|
LCE3B_HUMAN
|
||||||
| NC score | 0.117258 (rank : 52) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5TA77 | Gene names | LCE3B, LEP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3B (Late envelope protein 14). | |||||
|
KR414_HUMAN
|
||||||
| NC score | 0.116876 (rank : 53) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BYQ6 | Gene names | KRTAP4-14, KAP4.14, KRTAP4.14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-14 (Keratin-associated protein 4.14) (Ultrahigh sulfur keratin-associated protein 4.14). | |||||
|
KR102_HUMAN
|
||||||
| NC score | 0.116497 (rank : 54) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KRA24_HUMAN
|
||||||
| NC score | 0.116264 (rank : 55) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.116133 (rank : 56) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.115450 (rank : 57) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KRA15_HUMAN
|
||||||
| NC score | 0.115342 (rank : 58) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYS1, Q52LP6 | Gene names | KRTAP1-5, KAP1.5, KRTAP1.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-5 (Keratin-associated protein 1.5) (High sulfur keratin-associated protein 1.5). | |||||
|
KRA42_HUMAN
|
||||||
| NC score | 0.115012 (rank : 59) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BYR5 | Gene names | KRTAP4-2, KAP4.2, KRTAP4.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-2 (Keratin-associated protein 4.2) (Ultrahigh sulfur keratin-associated protein 4.2). | |||||
|
LSR_MOUSE
|
||||||
| NC score | 0.114832 (rank : 60) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99KG5, Q3TJE7, Q3UIQ9, Q61148, Q61149, Q6U816 | Gene names | Lsr, Lisch7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipolysis-stimulated lipoprotein receptor precursor (Liver-specific bHLH-Zip transcription factor) (Lipolysis-stimulated receptor) (Liver- specific gene on mouse chromosome 7 protein). | |||||
|
KR103_HUMAN
|
||||||
| NC score | 0.114627 (rank : 61) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
KR410_HUMAN
|
||||||
| NC score | 0.114568 (rank : 62) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
KRA47_HUMAN
|
||||||
| NC score | 0.114186 (rank : 63) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.112051 (rank : 64) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.111829 (rank : 65) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
LSR_HUMAN
|
||||||
| NC score | 0.111736 (rank : 66) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86X29, O00112, O00426, Q6ZT80, Q9UQL3 | Gene names | LSR, LISCH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipolysis-stimulated lipoprotein receptor. | |||||
|
KRA57_HUMAN
|
||||||
| NC score | 0.110498 (rank : 67) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
WFDC3_HUMAN
|
||||||
| NC score | 0.110348 (rank : 68) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IUB2, Q8TC52, Q9BQP3, Q9BQP4 | Gene names | WFDC3, WAP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 3 precursor (Putative protease inhibitor WAP14). | |||||
|
KRA98_HUMAN
|
||||||
| NC score | 0.109283 (rank : 69) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
KRA52_HUMAN
|
||||||
| NC score | 0.109252 (rank : 70) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA94_HUMAN
|
||||||
| NC score | 0.108740 (rank : 71) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYQ2 | Gene names | KRTAP9-4, KAP9.4, KRTAP9.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-4 (Keratin-associated protein 9.4) (Ultrahigh sulfur keratin-associated protein 9.4). | |||||
|
KRA44_HUMAN
|
||||||
| NC score | 0.107919 (rank : 72) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BYR3 | Gene names | KRTAP4-4, KAP4.4, KRTAP4.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-4 (Keratin-associated protein 4.4) (Ultrahigh sulfur keratin-associated protein 4.4). | |||||
|
KR108_HUMAN
|
||||||
| NC score | 0.107504 (rank : 73) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KRA92_HUMAN
|
||||||
| NC score | 0.105622 (rank : 74) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
KR109_HUMAN
|
||||||
| NC score | 0.104964 (rank : 75) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
KRA11_HUMAN
|
||||||
| NC score | 0.104189 (rank : 76) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q07627, Q96S60, Q96S67 | Gene names | KRTAP1-1, B2A, KAP1.1, KAP1.6, KAP1.7, KRTAP1.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-1 (Keratin-associated protein 1.1) (High sulfur keratin-associated protein 1.1) (Keratin-associated protein 1.6) (Keratin-associated protein 1.7). | |||||
|
SPR2A_MOUSE
|
||||||
| NC score | 0.103409 (rank : 77) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CQK8, O70553 | Gene names | Sprr2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A. | |||||
|
KRA13_HUMAN
|
||||||
| NC score | 0.103371 (rank : 78) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IUG1, Q07628, Q8IUG0, Q9BYS2 | Gene names | KRTAP1-3, B2B, KAP1.2, KAP1.3, KAP1.8, KAP1.9, KRATP1.9, KRTAP1.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-3 (Keratin-associated protein 1.8) (Keratin-associated protein 1.9). | |||||
|
LCE3A_HUMAN
|
||||||
| NC score | 0.100211 (rank : 79) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5TA76 | Gene names | LCE3A, LEP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3A (Late envelope protein 13). | |||||
|
KRA93_HUMAN
|
||||||
| NC score | 0.098454 (rank : 80) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.098142 (rank : 81) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.097147 (rank : 82) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KR413_HUMAN
|
||||||
| NC score | 0.094914 (rank : 83) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYU7 | Gene names | KRTAP4-13, KAP4.13, KRTAP4.13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-13 (Keratin-associated protein 4.13) (Ultrahigh sulfur keratin-associated protein 4.13). | |||||
|
KR10C_HUMAN
|
||||||
| NC score | 0.094723 (rank : 84) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KR122_HUMAN
|
||||||
| NC score | 0.094206 (rank : 85) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P59991 | Gene names | KRTAP12-2, KAP12.2, KRTAP12.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-2 (Keratin-associated protein 12.2) (High sulfur keratin-associated protein 12.2). | |||||
|
KR171_HUMAN
|
||||||
| NC score | 0.093480 (rank : 86) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
LCE3E_HUMAN
|
||||||
| NC score | 0.092234 (rank : 87) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5T5B0 | Gene names | LCE3E, LEP17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3E (Late envelope protein 17). | |||||
|
KR123_HUMAN
|
||||||
| NC score | 0.092051 (rank : 88) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P60328 | Gene names | KRTAP12-3, KAP12.3, KRTAP12.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-3 (Keratin-associated protein 12.3) (High sulfur keratin-associated protein 12.3). | |||||
|
KR10A_HUMAN
|
||||||
| NC score | 0.091666 (rank : 89) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P60014 | Gene names | KRTAP10-10, KAP10.10, KAP18-10, KRTAP10.10, KRTAP18-1, KRTAP18.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-10 (Keratin-associated protein 10.10) (High sulfur keratin-associated protein 10.10) (Keratin-associated protein 18-10) (Keratin-associated protein 18.10). | |||||
|
ADA10_MOUSE
|
||||||
| NC score | 0.090107 (rank : 90) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O35598 | Gene names | Adam10, Kuz, Madm | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 10 precursor (EC 3.4.24.81) (A disintegrin and metalloproteinase domain 10) (Mammalian disintegrin-metalloprotease) (Kuzbanian protein homolog). | |||||
|
ADA10_HUMAN
|
||||||
| NC score | 0.090069 (rank : 91) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14672, Q10742, Q92650 | Gene names | ADAM10, KUZ, MADM | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 10 precursor (EC 3.4.24.81) (A disintegrin and metalloproteinase domain 10) (Mammalian disintegrin-metalloprotease) (Kuzbanian protein homolog) (CDw156c antigen). | |||||
|
KRA99_HUMAN
|
||||||
| NC score | 0.089997 (rank : 92) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.089183 (rank : 93) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
KR121_HUMAN
|
||||||
| NC score | 0.085856 (rank : 94) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59990 | Gene names | KRTAP12-1, KAP12.1, KRTAP12.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-1 (Keratin-associated protein 12.1) (High sulfur keratin-associated protein 12.1). | |||||
|
WFDC8_HUMAN
|
||||||
| NC score | 0.083774 (rank : 95) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
KRA55_HUMAN
|
||||||
| NC score | 0.083599 (rank : 96) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.083141 (rank : 97) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.081538 (rank : 98) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.081280 (rank : 99) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
NFX1_HUMAN
|
||||||
| NC score | 0.077457 (rank : 100) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.077433 (rank : 101) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.076789 (rank : 102) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.076031 (rank : 103) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
MT3_HUMAN
|
||||||
| NC score | 0.075116 (rank : 104) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P25713, Q2V574 | Gene names | MT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-3 (MT-3) (Metallothionein-III) (MT-III) (Growth inhibitory factor) (GIF) (GIFB). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.071806 (rank : 105) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ALK1_MOUSE
|
||||||
| NC score | 0.071288 (rank : 106) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97430, O09081, O09082 | Gene names | Slpi | |||
|
Domain Architecture |

|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (Secretory leukocyte protease inhibitor). | |||||
|
ADA17_HUMAN
|
||||||
| NC score | 0.070612 (rank : 107) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P78536, O60226 | Gene names | ADAM17, CSVP, TACE | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (Snake venom-like protease) (CD156b antigen). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.070377 (rank : 108) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
LAMA5_MOUSE
|
||||||
| NC score | 0.069812 (rank : 109) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
ODFP_MOUSE
|
||||||
| NC score | 0.068973 (rank : 110) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61999 | Gene names | Odf1, Odfp | |||
|
Domain Architecture |

|
|||||
| Description | Outer dense fiber protein. | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.068615 (rank : 111) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
DLL3_HUMAN
|
||||||
| NC score | 0.068315 (rank : 112) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.065759 (rank : 113) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.065646 (rank : 114) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
LAMA5_HUMAN
|
||||||
| NC score | 0.063185 (rank : 115) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.061901 (rank : 116) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
PS1C2_HUMAN
|
||||||
| NC score | 0.061304 (rank : 117) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
ADA17_MOUSE
|
||||||
| NC score | 0.061260 (rank : 118) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z0F8, O88726, Q9R1U4, Q9Z0K3 | Gene names | Adam17, Tace | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (CD156b antigen). | |||||
|
KR111_HUMAN
|
||||||
| NC score | 0.061207 (rank : 119) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IUC1 | Gene names | KRTAP11-1, KAP11.1, KRTAP11.1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin-associated protein 11-1 (High sulfur keratin-associated protein 11.1). | |||||
|
WFDC2_HUMAN
|
||||||
| NC score | 0.060914 (rank : 120) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14508, Q8WXV9, Q8WXW0, Q8WXW1, Q8WXW2, Q96KJ1 | Gene names | WFDC2, HE4, WAP5 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 2 precursor (Major epididymis- specific protein E4) (Epididymal secretory protein E4) (Putative protease inhibitor WAP5). | |||||
|
EPPI_MOUSE
|
||||||
| NC score | 0.060896 (rank : 121) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DA01 | Gene names | Spinlw1 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1). | |||||
|
LRP1_HUMAN
|
||||||
| NC score | 0.060345 (rank : 122) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.058421 (rank : 123) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
WIF1_MOUSE
|
||||||
| NC score | 0.058357 (rank : 124) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
ALK1_HUMAN
|
||||||
| NC score | 0.058191 (rank : 125) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P03973, P07757 | Gene names | SLPI, WAP4, WFDC4 | |||
|
Domain Architecture |

|
|||||
| Description | Antileukoproteinase 1 precursor (ALP) (HUSI-1) (Seminal proteinase inhibitor) (Secretory leukocyte protease inhibitor) (BLPI) (Mucus proteinase inhibitor) (MPI) (WAP four-disulfide core domain protein 4) (Protease inhibitor WAP4). | |||||
|
ADA32_MOUSE
|
||||||
| NC score | 0.057847 (rank : 126) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K410, Q6P901, Q8BJ80 | Gene names | Adam32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAM 32 precursor (A disintegrin and metalloproteinase domain 32). | |||||
|
WIF1_HUMAN
|
||||||
| NC score | 0.056866 (rank : 127) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.056826 (rank : 128) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
NOTC4_MOUSE
|
||||||
| NC score | 0.056738 (rank : 129) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NAGPA_MOUSE
|
||||||
| NC score | 0.056714 (rank : 130) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BJ48, Q3UUT5, Q8CHQ8, Q9QZE6 | Gene names | Nagpa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.056508 (rank : 131) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
ZNFX1_MOUSE
|
||||||
| NC score | 0.056075 (rank : 132) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
DLL3_MOUSE
|
||||||
| NC score | 0.055995 (rank : 133) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
FRAS1_MOUSE
|
||||||
| NC score | 0.054927 (rank : 134) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
STAB1_MOUSE
|
||||||
| NC score | 0.054421 (rank : 135) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
STAB1_HUMAN
|
||||||
| NC score | 0.054281 (rank : 136) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
NICE1_HUMAN
|
||||||
| NC score | 0.053920 (rank : 137) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9UGL9 | Gene names | NICE1, C1orf42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein NICE-1. | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.053831 (rank : 138) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.053658 (rank : 139) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
DLL1_MOUSE
|
||||||
| NC score | 0.053551 (rank : 140) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
FBN2_HUMAN
|
||||||
| NC score | 0.053219 (rank : 141) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
LRP1B_HUMAN
|
||||||
| NC score | 0.053041 (rank : 142) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
DLK_HUMAN
|
||||||
| NC score | 0.053013 (rank : 143) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
ZNFX1_HUMAN
|
||||||
| NC score | 0.052680 (rank : 144) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
FBN1_HUMAN
|
||||||
| NC score | 0.052578 (rank : 145) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.052153 (rank : 146) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
FOLR3_HUMAN
|
||||||
| NC score | 0.052025 (rank : 147) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41439 | Gene names | FOLR3 | |||
|
Domain Architecture |

|
|||||
| Description | Folate receptor gamma precursor (FR-gamma) (Folate receptor 3). | |||||
|
FOLR2_HUMAN
|
||||||
| NC score | 0.052003 (rank : 148) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14207 | Gene names | FOLR2 | |||
|
Domain Architecture |

|
|||||
| Description | Folate receptor beta precursor (FR-beta) (Folate receptor 2) (Folate receptor, fetal/placental) (Placental folate-binding protein) (FBP). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.051459 (rank : 149) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
LRP1B_MOUSE
|
||||||
| NC score | 0.051458 (rank : 150) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.051289 (rank : 151) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
TENX_HUMAN
|
||||||
| NC score | 0.050801 (rank : 152) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
DLL4_MOUSE
|
||||||
| NC score | 0.050773 (rank : 153) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
FBN3_HUMAN
|
||||||
| NC score | 0.050606 (rank : 154) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
DLL1_HUMAN
|
||||||
| NC score | 0.050312 (rank : 155) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
FBN2_MOUSE
|
||||||
| NC score | 0.050247 (rank : 156) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
VWF_HUMAN
|
||||||
| NC score | 0.050243 (rank : 157) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.050062 (rank : 158) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.040008 (rank : 159) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.039189 (rank : 160) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
ERBB3_HUMAN
|
||||||
| NC score | 0.025720 (rank : 161) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P21860 | Gene names | ERBB3, HER3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Tyrosine kinase-type cell surface receptor HER3). | |||||
|
TINAL_MOUSE
|
||||||
| NC score | 0.024537 (rank : 162) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99JR5, Q9EQT9 | Gene names | Tinagl1, Arg1, Lcn7, Tinagl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Androgen- regulated gene 1 protein) (Adrenocortical zonation factor 1) (AZ-1) (Tubulointerstitial nephritis antigen-related protein) (TARP). | |||||
|
ERBB3_MOUSE
|
||||||
| NC score | 0.023378 (rank : 163) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 898 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61526, Q3KQR1, Q68J64, Q810U8, Q8K317 | Gene names | Erbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Glial growth factor receptor). | |||||
|
TNR3_HUMAN
|
||||||
| NC score | 0.022828 (rank : 164) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P36941 | Gene names | LTBR, TNFCR, TNFRSF3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor) (Tumor necrosis factor receptor 2-related protein) (Tumor necrosis factor C receptor). | |||||