Please be patient as the page loads
|
AMBP_MOUSE
|
||||||
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |
|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AMBP_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
AMBP_HUMAN
|
||||||
| θ value | 3.37801e-156 (rank : 2) | NC score | 0.990889 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02760, P00977, P02759 | Gene names | AMBP, HCP, ITIL | |||
|
Domain Architecture |
|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin (Protein HC) (Complex-forming glycoprotein heterogeneous in charge) (Alpha-1 microglycoprotein); Inter-alpha-trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
SPIT2_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 3) | NC score | 0.717763 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WU03, Q9WU04, Q9WU05 | Gene names | Spint2, Hai2 | |||
|
Domain Architecture |
|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2). | |||||
|
SPIT2_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 4) | NC score | 0.710150 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43291, O00271, O14895, Q5TZQ3, Q969E0 | Gene names | SPINT2, HAI2, KOP | |||
|
Domain Architecture |
|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2) (Placental bikunin). | |||||
|
TFPI1_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 5) | NC score | 0.696319 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P10646, O95103 | Gene names | TFPI, LACI, TFPI1 | |||
|
Domain Architecture |
|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
TFPI1_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 6) | NC score | 0.687441 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54819, Q9Z2U8 | Gene names | Tfpi | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
PTGDS_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 7) | NC score | 0.552411 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P41222, Q5SQ10, Q7M4P3, Q9UCC9, Q9UCD9 | Gene names | PTGDS, PDS | |||
|
Domain Architecture |
|
|||||
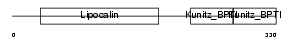
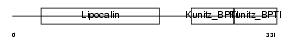
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS) (Beta-trace protein) (Cerebrin-28). | |||||
|
CO8G_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 8) | NC score | 0.521451 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VCG4 | Gene names | C8g | |||
|
Domain Architecture |
|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
PTGDS_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 9) | NC score | 0.544524 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O09114, O09157, O35091, Q62169 | Gene names | Ptgds | |||
|
Domain Architecture |
|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS). | |||||
|
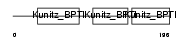
TFPI2_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 10) | NC score | 0.674093 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P48307 | Gene names | TFPI2 | |||
|
Domain Architecture |
|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2) (Placental protein 5) (PP5). | |||||
|
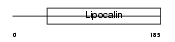
CO8G_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 11) | NC score | 0.493795 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P07360, Q5SQ07 | Gene names | C8G | |||
|
Domain Architecture |
|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
SPIT1_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 12) | NC score | 0.535442 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R097 | Gene names | Spint1, Hai1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
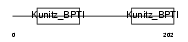
TFPI2_MOUSE
|
||||||
| θ value | 2.98157e-11 (rank : 13) | NC score | 0.661582 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35536 | Gene names | Tfpi2 | |||
|
Domain Architecture |
|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2). | |||||
|
A4_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 14) | NC score | 0.442529 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
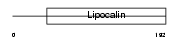
NGAL_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 15) | NC score | 0.435349 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P80188, P30150, Q92683 | Gene names | LCN2, HNL, NGAL | |||
|
Domain Architecture |
|
|||||
| Description | Neutrophil gelatinase-associated lipocalin precursor (NGAL) (p25) (25 kDa alpha-2-microglobulin-related subunit of MMP-9) (Lipocalin-2) (Oncogene 24p3). | |||||
|
SPIT1_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 16) | NC score | 0.510951 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
APLP2_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 17) | NC score | 0.438555 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
A4_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 18) | NC score | 0.439690 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
CO6A3_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 19) | NC score | 0.222305 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
EPPI_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 20) | NC score | 0.601537 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95925, Q86TP9, Q96SD7, Q9HD30 | Gene names | SPINLW1, WAP7, WFDC7 | |||
|
Domain Architecture |

|
|||||
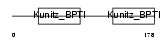
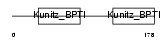
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1) (WAP four-disulfide core domain protein 7) (Protease inhibitor WAP7). | |||||
|
EPPI_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 21) | NC score | 0.591540 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DA01 | Gene names | Spinlw1 | |||
|
Domain Architecture |
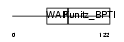
|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1). | |||||
|
WFDC8_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 22) | NC score | 0.499580 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |
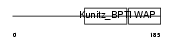
|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
OBP2B_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 23) | NC score | 0.364800 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NPH6, Q9NY51, Q9NY52 | Gene names | OBP2B | |||
|
Domain Architecture |
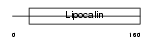
|
|||||
| Description | Odorant-binding protein 2b precursor (OBPIIb). | |||||
|
MUP5_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 24) | NC score | 0.337628 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11591 | Gene names | Mup5 | |||
|
Domain Architecture |
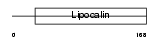
|
|||||
| Description | Major urinary protein 5 precursor (MUP 5). | |||||
|
VNS1_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 25) | NC score | 0.383913 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62471 | Gene names | Lcn3 | |||
|
Domain Architecture |
|
|||||
| Description | Vomeronasal secretory protein 1 precursor (Vomeronasal secretory protein I) (VNSP I) (Lipocalin-3). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 26) | NC score | 0.109586 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
LCN1_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 27) | NC score | 0.364124 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P31025 | Gene names | LCN1, VEGP | |||
|
Domain Architecture |
|
|||||
| Description | Lipocalin-1 precursor (Von Ebner gland protein) (VEG protein) (Tear prealbumin) (TP) (Tear lipocalin) (Tlc). | |||||
|
OBP2A_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 28) | NC score | 0.297972 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NY56, Q9NY50, Q9NY53, Q9NY54, Q9NY55 | Gene names | OBP2A | |||
|
Domain Architecture |
|
|||||
| Description | Odorant-binding protein 2a precursor (OBPIIa). | |||||
|
NGAL_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 29) | NC score | 0.371055 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P11672 | Gene names | Lcn2 | |||
|
Domain Architecture |
|
|||||
| Description | Neutrophil gelatinase-associated lipocalin precursor (NGAL) (p25) (SV- 40-induced 24P3 protein) (Lipocalin-2). | |||||
|
LCN13_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 30) | NC score | 0.324675 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K1H9 | Gene names | Lcn13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-13 precursor. | |||||
|
SPIT3_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 31) | NC score | 0.665456 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49223, Q6UDR8, Q96KK2 | Gene names | SPINT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kunitz-type protease inhibitor 3 precursor (HKIB9). | |||||
|
MUP1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 32) | NC score | 0.314123 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11588, Q61921 | Gene names | Mup1 | |||
|
Domain Architecture |
|
|||||
| Description | Major urinary protein 1 precursor (MUP 1). | |||||
|
MUP6_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 33) | NC score | 0.313503 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P02762, P70119, Q78EF5, Q91V46 | Gene names | Mup6 | |||
|
Domain Architecture |
|
|||||
| Description | Major urinary protein 6 precursor (MUP 6) (Alpha-2U-globulin) (Group 1, BS6) (Allergen Mus m 1). | |||||
|
LCN9_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 34) | NC score | 0.288913 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WX39, Q6JVE7 | Gene names | LCN9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-9 precursor (MUP-like lipocalin). | |||||
|
MUP2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 35) | NC score | 0.313925 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11589, Q99JF6 | Gene names | Mup2 | |||
|
Domain Architecture |
|
|||||
| Description | Major urinary protein 2 precursor (MUP 2). | |||||
|
MUP3_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 36) | NC score | 0.320675 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P04939, P97897, Q8VCG6 | Gene names | Mup3 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 3 precursor (MUP 3) (Non-group 1/group 2 MUP15). | |||||
|
MUP4_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 37) | NC score | 0.323109 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11590 | Gene names | Mup4 | |||
|
Domain Architecture |
|
|||||
| Description | Major urinary protein 4 precursor (MUP 4). | |||||
|
LCN9_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 38) | NC score | 0.333415 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D267, Q80ZC4 | Gene names | Lcn9 | |||
|
Domain Architecture |
|
|||||
| Description | Epididymal-specific lipocalin-9 precursor (MUP-like lipocalin). | |||||
|
RETBP_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 39) | NC score | 0.148663 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02753, O43478, O43479, Q5VY24, Q8WWA3, Q9P178 | Gene names | RBP4 | |||
|
Domain Architecture |
|
|||||
| Description | Plasma retinol-binding protein precursor (PRBP) (RBP) [Contains: Plasma retinol-binding protein(1-182); Plasma retinol-binding protein(1-181); Plasma retinol-binding protein(1-179); Plasma retinol- binding protein(1-176)]. | |||||
|
WFDC6_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 40) | NC score | 0.380134 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BQY6, Q5JYQ4, Q8NFV6 | Gene names | WFDC6, C20orf171, WAP6 | |||
|
Domain Architecture |
|
|||||
| Description | WAP four-disulfide core domain protein 6 precursor (Putative protease inhibitor WAP6). | |||||
|
RETBP_MOUSE
|
||||||
| θ value | 0.21417 (rank : 41) | NC score | 0.131168 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00724, P70357, Q566I5 | Gene names | Rbp4 | |||
|
Domain Architecture |
|
|||||
| Description | Plasma retinol-binding protein precursor (PRBP) (RBP). | |||||
|
KITM_MOUSE
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.050191 (rank : 55) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R088 | Gene names | Tk2 | |||
|
Domain Architecture |
|
|||||
| Description | Thymidine kinase 2, mitochondrial precursor (EC 2.7.1.21) (Mt-TK). | |||||
|
KITM_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.047617 (rank : 56) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00142, O15238 | Gene names | TK2 | |||
|
Domain Architecture |

|
|||||
| Description | Thymidine kinase 2, mitochondrial precursor (EC 2.7.1.21) (Mt-TK). | |||||
|
CAPS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.030224 (rank : 58) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULU8, Q13339, Q6GQQ6, Q8N2Z5, Q8N3M7, Q8NFR0, Q96BC2 | Gene names | CADPS, CAPS, CAPS1, KIAA1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
CAPS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.032809 (rank : 57) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80TJ1, Q3TSP2, Q61374, Q6AXB4, Q6PGF0 | Gene names | Cadps, Caps, Caps1, Kiaa1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
CS016_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.015385 (rank : 59) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
STAB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.002041 (rank : 60) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
VNS2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.205083 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62472 | Gene names | Lcn4 | |||
|
Domain Architecture |
|
|||||
| Description | Vomeronasal secretory protein 2 precursor (Vomeronasal secretory protein II) (VNSP II) (Lipocalin-4). | |||||
|
APLP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.120756 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.120491 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.143550 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06335 | Gene names | Aplp2 | |||
|
Domain Architecture |
|
|||||
| Description | Amyloid-like protein 2 precursor (CDEI box-binding protein) (CDEBP). | |||||
|
LCN12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.149455 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6JVE5 | Gene names | LCN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-12 precursor. | |||||
|
LCN12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.158427 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6JVL5 | Gene names | Lcn12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-12 precursor. | |||||
|
LCN6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.089698 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62502, Q71SF6 | Gene names | LCN6, LCN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-6 precursor (Lipocalin-5). | |||||
|
LRP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.062790 (rank : 53) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86VZ4, Q5VYC0, Q96SN6 | Gene names | LRP11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
LRP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.063247 (rank : 52) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CB67, Q8C7Y7 | Gene names | Lrp11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
PAEP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.172428 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P09466, Q9UG92 | Gene names | PAEP | |||
|
Domain Architecture |
|
|||||
| Description | Glycodelin precursor (GD) (Pregnancy-associated endometrial alpha-2 globulin) (PEG) (PAEG) (Placental protein 14) (PP14) (Progesterone- associated endometrial protein) (Progestagen-associated endometrial protein). | |||||
|
PBAS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.113700 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08976 | Gene names | Pbsn, Prbs | |||
|
Domain Architecture |
|
|||||
| Description | Probasin precursor (PB). | |||||
|
WFD12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.050861 (rank : 54) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHY3 | Gene names | Wfdc12, Swam2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 12 precursor (Whey acidic protein 2) (Single WAP motif protein 2) (Elafin-like protein II). | |||||
|
WFD15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.084488 (rank : 51) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHY4, Q8BVC0 | Gene names | Wfdc15, Swam1 | |||
|
Domain Architecture |
|
|||||
| Description | WAP four-disulfide core domain protein 15 precursor (Single WAP motif protein 1) (Elafin-like protein I). | |||||
|
AMBP_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
AMBP_HUMAN
|
||||||
| NC score | 0.990889 (rank : 2) | θ value | 3.37801e-156 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02760, P00977, P02759 | Gene names | AMBP, HCP, ITIL | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin (Protein HC) (Complex-forming glycoprotein heterogeneous in charge) (Alpha-1 microglycoprotein); Inter-alpha-trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
SPIT2_MOUSE
|
||||||
| NC score | 0.717763 (rank : 3) | θ value | 1.24977e-17 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WU03, Q9WU04, Q9WU05 | Gene names | Spint2, Hai2 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2). | |||||
|
SPIT2_HUMAN
|
||||||
| NC score | 0.710150 (rank : 4) | θ value | 3.63628e-17 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43291, O00271, O14895, Q5TZQ3, Q969E0 | Gene names | SPINT2, HAI2, KOP | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 2 precursor (Hepatocyte growth factor activator inhibitor type 2) (HAI-2) (Placental bikunin). | |||||
|
TFPI1_HUMAN
|
||||||
| NC score | 0.696319 (rank : 5) | θ value | 4.02038e-16 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P10646, O95103 | Gene names | TFPI, LACI, TFPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
TFPI1_MOUSE
|
||||||
| NC score | 0.687441 (rank : 6) | θ value | 6.85773e-16 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54819, Q9Z2U8 | Gene names | Tfpi | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor precursor (TFPI) (Lipoprotein- associated coagulation inhibitor) (LACI) (Extrinsic pathway inhibitor) (EPI). | |||||
|
TFPI2_HUMAN
|
||||||
| NC score | 0.674093 (rank : 7) | θ value | 9.26847e-13 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P48307 | Gene names | TFPI2 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2) (Placental protein 5) (PP5). | |||||
|
SPIT3_HUMAN
|
||||||
| NC score | 0.665456 (rank : 8) | θ value | 0.000461057 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49223, Q6UDR8, Q96KK2 | Gene names | SPINT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kunitz-type protease inhibitor 3 precursor (HKIB9). | |||||
|
TFPI2_MOUSE
|
||||||
| NC score | 0.661582 (rank : 9) | θ value | 2.98157e-11 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35536 | Gene names | Tfpi2 | |||
|
Domain Architecture |

|
|||||
| Description | Tissue factor pathway inhibitor 2 precursor (TFPI-2). | |||||
|
EPPI_HUMAN
|
||||||
| NC score | 0.601537 (rank : 10) | θ value | 6.21693e-09 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95925, Q86TP9, Q96SD7, Q9HD30 | Gene names | SPINLW1, WAP7, WFDC7 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1) (WAP four-disulfide core domain protein 7) (Protease inhibitor WAP7). | |||||
|
EPPI_MOUSE
|
||||||
| NC score | 0.591540 (rank : 11) | θ value | 4.45536e-07 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DA01 | Gene names | Spinlw1 | |||
|
Domain Architecture |

|
|||||
| Description | Eppin precursor (Epididymal protease inhibitor) (Serine protease inhibitor-like with Kunitz and WAP domains 1). | |||||
|
PTGDS_HUMAN
|
||||||
| NC score | 0.552411 (rank : 12) | θ value | 2.60593e-15 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P41222, Q5SQ10, Q7M4P3, Q9UCC9, Q9UCD9 | Gene names | PTGDS, PDS | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS) (Beta-trace protein) (Cerebrin-28). | |||||
|
PTGDS_MOUSE
|
||||||
| NC score | 0.544524 (rank : 13) | θ value | 1.86753e-13 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O09114, O09157, O35091, Q62169 | Gene names | Ptgds | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS). | |||||
|
SPIT1_MOUSE
|
||||||
| NC score | 0.535442 (rank : 14) | θ value | 1.33837e-11 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R097 | Gene names | Spint1, Hai1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
CO8G_MOUSE
|
||||||
| NC score | 0.521451 (rank : 15) | θ value | 1.68911e-14 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VCG4 | Gene names | C8g | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
SPIT1_HUMAN
|
||||||
| NC score | 0.510951 (rank : 16) | θ value | 3.29651e-10 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
WFDC8_HUMAN
|
||||||
| NC score | 0.499580 (rank : 17) | θ value | 9.92553e-07 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
CO8G_HUMAN
|
||||||
| NC score | 0.493795 (rank : 18) | θ value | 1.2105e-12 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P07360, Q5SQ07 | Gene names | C8G | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.442529 (rank : 19) | θ value | 2.52405e-10 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_HUMAN
|
||||||
| NC score | 0.439690 (rank : 20) | θ value | 7.34386e-10 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
APLP2_HUMAN
|
||||||
| NC score | 0.438555 (rank : 21) | θ value | 4.30538e-10 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
NGAL_HUMAN
|
||||||
| NC score | 0.435349 (rank : 22) | θ value | 2.52405e-10 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P80188, P30150, Q92683 | Gene names | LCN2, HNL, NGAL | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil gelatinase-associated lipocalin precursor (NGAL) (p25) (25 kDa alpha-2-microglobulin-related subunit of MMP-9) (Lipocalin-2) (Oncogene 24p3). | |||||
|
VNS1_MOUSE
|
||||||
| NC score | 0.383913 (rank : 23) | θ value | 1.87187e-05 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62471 | Gene names | Lcn3 | |||
|
Domain Architecture |

|
|||||
| Description | Vomeronasal secretory protein 1 precursor (Vomeronasal secretory protein I) (VNSP I) (Lipocalin-3). | |||||
|
WFDC6_HUMAN
|
||||||
| NC score | 0.380134 (rank : 24) | θ value | 0.0563607 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BQY6, Q5JYQ4, Q8NFV6 | Gene names | WFDC6, C20orf171, WAP6 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 6 precursor (Putative protease inhibitor WAP6). | |||||
|
NGAL_MOUSE
|
||||||
| NC score | 0.371055 (rank : 25) | θ value | 0.000121331 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P11672 | Gene names | Lcn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil gelatinase-associated lipocalin precursor (NGAL) (p25) (SV- 40-induced 24P3 protein) (Lipocalin-2). | |||||
|
OBP2B_HUMAN
|
||||||
| NC score | 0.364800 (rank : 26) | θ value | 3.77169e-06 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NPH6, Q9NY51, Q9NY52 | Gene names | OBP2B | |||
|
Domain Architecture |

|
|||||
| Description | Odorant-binding protein 2b precursor (OBPIIb). | |||||
|
LCN1_HUMAN
|
||||||
| NC score | 0.364124 (rank : 27) | θ value | 2.44474e-05 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P31025 | Gene names | LCN1, VEGP | |||
|
Domain Architecture |

|
|||||
| Description | Lipocalin-1 precursor (Von Ebner gland protein) (VEG protein) (Tear prealbumin) (TP) (Tear lipocalin) (Tlc). | |||||
|
MUP5_MOUSE
|
||||||
| NC score | 0.337628 (rank : 28) | θ value | 1.87187e-05 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11591 | Gene names | Mup5 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 5 precursor (MUP 5). | |||||
|
LCN9_MOUSE
|
||||||
| NC score | 0.333415 (rank : 29) | θ value | 0.00509761 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D267, Q80ZC4 | Gene names | Lcn9 | |||
|
Domain Architecture |

|
|||||
| Description | Epididymal-specific lipocalin-9 precursor (MUP-like lipocalin). | |||||
|
LCN13_MOUSE
|
||||||
| NC score | 0.324675 (rank : 30) | θ value | 0.000461057 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K1H9 | Gene names | Lcn13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-13 precursor. | |||||
|
MUP4_MOUSE
|
||||||
| NC score | 0.323109 (rank : 31) | θ value | 0.00228821 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11590 | Gene names | Mup4 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 4 precursor (MUP 4). | |||||
|
MUP3_MOUSE
|
||||||
| NC score | 0.320675 (rank : 32) | θ value | 0.00175202 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P04939, P97897, Q8VCG6 | Gene names | Mup3 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 3 precursor (MUP 3) (Non-group 1/group 2 MUP15). | |||||
|
MUP1_MOUSE
|
||||||
| NC score | 0.314123 (rank : 33) | θ value | 0.00102713 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11588, Q61921 | Gene names | Mup1 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 1 precursor (MUP 1). | |||||
|
MUP2_MOUSE
|
||||||
| NC score | 0.313925 (rank : 34) | θ value | 0.00175202 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11589, Q99JF6 | Gene names | Mup2 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 2 precursor (MUP 2). | |||||
|
MUP6_MOUSE
|
||||||
| NC score | 0.313503 (rank : 35) | θ value | 0.00102713 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P02762, P70119, Q78EF5, Q91V46 | Gene names | Mup6 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 6 precursor (MUP 6) (Alpha-2U-globulin) (Group 1, BS6) (Allergen Mus m 1). | |||||
|
OBP2A_HUMAN
|
||||||
| NC score | 0.297972 (rank : 36) | θ value | 2.44474e-05 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NY56, Q9NY50, Q9NY53, Q9NY54, Q9NY55 | Gene names | OBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Odorant-binding protein 2a precursor (OBPIIa). | |||||
|
LCN9_HUMAN
|
||||||
| NC score | 0.288913 (rank : 37) | θ value | 0.00175202 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WX39, Q6JVE7 | Gene names | LCN9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-9 precursor (MUP-like lipocalin). | |||||
|
CO6A3_HUMAN
|
||||||
| NC score | 0.222305 (rank : 38) | θ value | 1.63604e-09 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
VNS2_MOUSE
|
||||||
| NC score | 0.205083 (rank : 39) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62472 | Gene names | Lcn4 | |||
|
Domain Architecture |

|
|||||
| Description | Vomeronasal secretory protein 2 precursor (Vomeronasal secretory protein II) (VNSP II) (Lipocalin-4). | |||||
|
PAEP_HUMAN
|
||||||
| NC score | 0.172428 (rank : 40) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P09466, Q9UG92 | Gene names | PAEP | |||
|
Domain Architecture |

|
|||||
| Description | Glycodelin precursor (GD) (Pregnancy-associated endometrial alpha-2 globulin) (PEG) (PAEG) (Placental protein 14) (PP14) (Progesterone- associated endometrial protein) (Progestagen-associated endometrial protein). | |||||
|
LCN12_MOUSE
|
||||||
| NC score | 0.158427 (rank : 41) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6JVL5 | Gene names | Lcn12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-12 precursor. | |||||
|
LCN12_HUMAN
|
||||||
| NC score | 0.149455 (rank : 42) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6JVE5 | Gene names | LCN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-12 precursor. | |||||
|
RETBP_HUMAN
|
||||||
| NC score | 0.148663 (rank : 43) | θ value | 0.0113563 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02753, O43478, O43479, Q5VY24, Q8WWA3, Q9P178 | Gene names | RBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma retinol-binding protein precursor (PRBP) (RBP) [Contains: Plasma retinol-binding protein(1-182); Plasma retinol-binding protein(1-181); Plasma retinol-binding protein(1-179); Plasma retinol- binding protein(1-176)]. | |||||
|
APLP2_MOUSE
|
||||||
| NC score | 0.143550 (rank : 44) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06335 | Gene names | Aplp2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (CDEI box-binding protein) (CDEBP). | |||||
|
RETBP_MOUSE
|
||||||
| NC score | 0.131168 (rank : 45) | θ value | 0.21417 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00724, P70357, Q566I5 | Gene names | Rbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma retinol-binding protein precursor (PRBP) (RBP). | |||||
|
APLP1_HUMAN
|
||||||
| NC score | 0.120756 (rank : 46) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
APLP1_MOUSE
|
||||||
| NC score | 0.120491 (rank : 47) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
PBAS_MOUSE
|
||||||
| NC score | 0.113700 (rank : 48) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08976 | Gene names | Pbsn, Prbs | |||
|
Domain Architecture |

|
|||||
| Description | Probasin precursor (PB). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.109586 (rank : 49) | θ value | 2.44474e-05 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
LCN6_HUMAN
|
||||||
| NC score | 0.089698 (rank : 50) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62502, Q71SF6 | Gene names | LCN6, LCN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-6 precursor (Lipocalin-5). | |||||
|
WFD15_MOUSE
|
||||||
| NC score | 0.084488 (rank : 51) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHY4, Q8BVC0 | Gene names | Wfdc15, Swam1 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 15 precursor (Single WAP motif protein 1) (Elafin-like protein I). | |||||
|
LRP11_MOUSE
|
||||||
| NC score | 0.063247 (rank : 52) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CB67, Q8C7Y7 | Gene names | Lrp11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
LRP11_HUMAN
|
||||||
| NC score | 0.062790 (rank : 53) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86VZ4, Q5VYC0, Q96SN6 | Gene names | LRP11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
WFD12_MOUSE
|
||||||
| NC score | 0.050861 (rank : 54) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHY3 | Gene names | Wfdc12, Swam2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 12 precursor (Whey acidic protein 2) (Single WAP motif protein 2) (Elafin-like protein II). | |||||
|
KITM_MOUSE
|
||||||
| NC score | 0.050191 (rank : 55) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R088 | Gene names | Tk2 | |||
|
Domain Architecture |

|
|||||
| Description | Thymidine kinase 2, mitochondrial precursor (EC 2.7.1.21) (Mt-TK). | |||||
|
KITM_HUMAN
|
||||||
| NC score | 0.047617 (rank : 56) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00142, O15238 | Gene names | TK2 | |||
|
Domain Architecture |

|
|||||
| Description | Thymidine kinase 2, mitochondrial precursor (EC 2.7.1.21) (Mt-TK). | |||||
|
CAPS1_MOUSE
|
||||||
| NC score | 0.032809 (rank : 57) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80TJ1, Q3TSP2, Q61374, Q6AXB4, Q6PGF0 | Gene names | Cadps, Caps, Caps1, Kiaa1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
CAPS1_HUMAN
|
||||||
| NC score | 0.030224 (rank : 58) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULU8, Q13339, Q6GQQ6, Q8N2Z5, Q8N3M7, Q8NFR0, Q96BC2 | Gene names | CADPS, CAPS, CAPS1, KIAA1121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 1 (Calcium-dependent activator protein for secretion 1) (CAPS-1). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.015385 (rank : 59) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
STAB1_HUMAN
|
||||||
| NC score | 0.002041 (rank : 60) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||