Please be patient as the page loads
|
KITM_MOUSE
|
||||||
| SwissProt Accessions | Q9R088 | Gene names | Tk2 | |||
|
Domain Architecture |
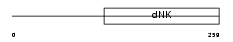
|
|||||
| Description | Thymidine kinase 2, mitochondrial precursor (EC 2.7.1.21) (Mt-TK). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KITM_MOUSE
|
||||||
| θ value | 3.87392e-144 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R088 | Gene names | Tk2 | |||
|
Domain Architecture |

|
|||||
| Description | Thymidine kinase 2, mitochondrial precursor (EC 2.7.1.21) (Mt-TK). | |||||
|
KITM_HUMAN
|
||||||
| θ value | 1.8752e-114 (rank : 2) | NC score | 0.997615 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00142, O15238 | Gene names | TK2 | |||
|
Domain Architecture |

|
|||||
| Description | Thymidine kinase 2, mitochondrial precursor (EC 2.7.1.21) (Mt-TK). | |||||
|
DCK_MOUSE
|
||||||
| θ value | 4.72714e-33 (rank : 3) | NC score | 0.875677 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P43346 | Gene names | Dck | |||
|
Domain Architecture |

|
|||||
| Description | Deoxycytidine kinase (EC 2.7.1.74) (dCK). | |||||
|
DCK_HUMAN
|
||||||
| θ value | 1.37539e-32 (rank : 4) | NC score | 0.874290 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27707 | Gene names | DCK | |||
|
Domain Architecture |

|
|||||
| Description | Deoxycytidine kinase (EC 2.7.1.74) (dCK). | |||||
|
DGUOK_MOUSE
|
||||||
| θ value | 1.02001e-27 (rank : 5) | NC score | 0.852605 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QX60, Q9R0Y2, Q9WUT9 | Gene names | Dguok, Dgk | |||
|
Domain Architecture |
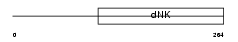
|
|||||
| Description | Deoxyguanosine kinase, mitochondrial precursor (EC 2.7.1.113) (dGK). | |||||
|
DGUOK_HUMAN
|
||||||
| θ value | 2.27234e-27 (rank : 6) | NC score | 0.850019 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16854, P78532, Q16759, Q96BC1 | Gene names | DGUOK, DGK | |||
|
Domain Architecture |
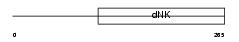
|
|||||
| Description | Deoxyguanosine kinase, mitochondrial precursor (EC 2.7.1.113) (dGK). | |||||
|
AMBP_MOUSE
|
||||||
| θ value | 0.47712 (rank : 7) | NC score | 0.050191 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |
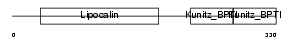
|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
CHSS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 8) | NC score | 0.027515 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
CHSS2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 9) | NC score | 0.027321 (rank : 10) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
NDUAA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 10) | NC score | 0.106152 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LC3, Q8BL57, Q9CW21 | Gene names | Ndufa10 | |||
|
Domain Architecture |
|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 42 kDa subunit) (Complex I-42kD) (CI-42kD). | |||||
|
KITM_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.87392e-144 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R088 | Gene names | Tk2 | |||
|
Domain Architecture |

|
|||||
| Description | Thymidine kinase 2, mitochondrial precursor (EC 2.7.1.21) (Mt-TK). | |||||
|
KITM_HUMAN
|
||||||
| NC score | 0.997615 (rank : 2) | θ value | 1.8752e-114 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00142, O15238 | Gene names | TK2 | |||
|
Domain Architecture |

|
|||||
| Description | Thymidine kinase 2, mitochondrial precursor (EC 2.7.1.21) (Mt-TK). | |||||
|
DCK_MOUSE
|
||||||
| NC score | 0.875677 (rank : 3) | θ value | 4.72714e-33 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P43346 | Gene names | Dck | |||
|
Domain Architecture |

|
|||||
| Description | Deoxycytidine kinase (EC 2.7.1.74) (dCK). | |||||
|
DCK_HUMAN
|
||||||
| NC score | 0.874290 (rank : 4) | θ value | 1.37539e-32 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27707 | Gene names | DCK | |||
|
Domain Architecture |

|
|||||
| Description | Deoxycytidine kinase (EC 2.7.1.74) (dCK). | |||||
|
DGUOK_MOUSE
|
||||||
| NC score | 0.852605 (rank : 5) | θ value | 1.02001e-27 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QX60, Q9R0Y2, Q9WUT9 | Gene names | Dguok, Dgk | |||
|
Domain Architecture |

|
|||||
| Description | Deoxyguanosine kinase, mitochondrial precursor (EC 2.7.1.113) (dGK). | |||||
|
DGUOK_HUMAN
|
||||||
| NC score | 0.850019 (rank : 6) | θ value | 2.27234e-27 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16854, P78532, Q16759, Q96BC1 | Gene names | DGUOK, DGK | |||
|
Domain Architecture |

|
|||||
| Description | Deoxyguanosine kinase, mitochondrial precursor (EC 2.7.1.113) (dGK). | |||||
|
NDUAA_MOUSE
|
||||||
| NC score | 0.106152 (rank : 7) | θ value | θ > 10 (rank : 10) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LC3, Q8BL57, Q9CW21 | Gene names | Ndufa10 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 42 kDa subunit) (Complex I-42kD) (CI-42kD). | |||||
|
AMBP_MOUSE
|
||||||
| NC score | 0.050191 (rank : 8) | θ value | 0.47712 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
CHSS2_HUMAN
|
||||||
| NC score | 0.027515 (rank : 9) | θ value | 4.03905 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
CHSS2_MOUSE
|
||||||
| NC score | 0.027321 (rank : 10) | θ value | 4.03905 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||