Please be patient as the page loads
|
MUP5_MOUSE
|
||||||
| SwissProt Accessions | P11591 | Gene names | Mup5 | |||
|
Domain Architecture |
|
|||||
| Description | Major urinary protein 5 precursor (MUP 5). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MUP5_MOUSE
|
||||||
| θ value | 2.38316e-93 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P11591 | Gene names | Mup5 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 5 precursor (MUP 5). | |||||
|
MUP4_MOUSE
|
||||||
| θ value | 2.55803e-79 (rank : 2) | NC score | 0.996747 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P11590 | Gene names | Mup4 | |||
|
Domain Architecture |
|
|||||
| Description | Major urinary protein 4 precursor (MUP 4). | |||||
|
MUP6_MOUSE
|
||||||
| θ value | 2.47765e-74 (rank : 3) | NC score | 0.994550 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P02762, P70119, Q78EF5, Q91V46 | Gene names | Mup6 | |||
|
Domain Architecture |
|
|||||
| Description | Major urinary protein 6 precursor (MUP 6) (Alpha-2U-globulin) (Group 1, BS6) (Allergen Mus m 1). | |||||
|
MUP2_MOUSE
|
||||||
| θ value | 4.22625e-74 (rank : 4) | NC score | 0.994344 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11589, Q99JF6 | Gene names | Mup2 | |||
|
Domain Architecture |
|
|||||
| Description | Major urinary protein 2 precursor (MUP 2). | |||||
|
MUP1_MOUSE
|
||||||
| θ value | 7.20884e-74 (rank : 5) | NC score | 0.994533 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11588, Q61921 | Gene names | Mup1 | |||
|
Domain Architecture |
|
|||||
| Description | Major urinary protein 1 precursor (MUP 1). | |||||
|
MUP3_MOUSE
|
||||||
| θ value | 2.83479e-70 (rank : 6) | NC score | 0.994217 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P04939, P97897, Q8VCG6 | Gene names | Mup3 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 3 precursor (MUP 3) (Non-group 1/group 2 MUP15). | |||||
|
LCN9_HUMAN
|
||||||
| θ value | 5.40856e-29 (rank : 7) | NC score | 0.918141 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WX39, Q6JVE7 | Gene names | LCN9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-9 precursor (MUP-like lipocalin). | |||||
|
LCN9_MOUSE
|
||||||
| θ value | 5.06226e-27 (rank : 8) | NC score | 0.909736 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D267, Q80ZC4 | Gene names | Lcn9 | |||
|
Domain Architecture |
|
|||||
| Description | Epididymal-specific lipocalin-9 precursor (MUP-like lipocalin). | |||||
|
VNS1_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 9) | NC score | 0.601079 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62471 | Gene names | Lcn3 | |||
|
Domain Architecture |
|
|||||
| Description | Vomeronasal secretory protein 1 precursor (Vomeronasal secretory protein I) (VNSP I) (Lipocalin-3). | |||||
|
PTGDS_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 10) | NC score | 0.554308 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41222, Q5SQ10, Q7M4P3, Q9UCC9, Q9UCD9 | Gene names | PTGDS, PDS | |||
|
Domain Architecture |
|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS) (Beta-trace protein) (Cerebrin-28). | |||||
|
PTGDS_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 11) | NC score | 0.525906 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O09114, O09157, O35091, Q62169 | Gene names | Ptgds | |||
|
Domain Architecture |
|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS). | |||||
|
NGAL_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 12) | NC score | 0.507710 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P11672 | Gene names | Lcn2 | |||
|
Domain Architecture |
|
|||||
| Description | Neutrophil gelatinase-associated lipocalin precursor (NGAL) (p25) (SV- 40-induced 24P3 protein) (Lipocalin-2). | |||||
|
PBAS_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 13) | NC score | 0.605885 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08976 | Gene names | Pbsn, Prbs | |||
|
Domain Architecture |
|
|||||
| Description | Probasin precursor (PB). | |||||
|
AMBP_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 14) | NC score | 0.337628 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |
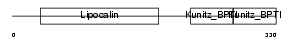
|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
PAEP_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 15) | NC score | 0.568318 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P09466, Q9UG92 | Gene names | PAEP | |||
|
Domain Architecture |
|
|||||
| Description | Glycodelin precursor (GD) (Pregnancy-associated endometrial alpha-2 globulin) (PEG) (PAEG) (Placental protein 14) (PP14) (Progesterone- associated endometrial protein) (Progestagen-associated endometrial protein). | |||||
|
AMBP_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 16) | NC score | 0.334853 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P02760, P00977, P02759 | Gene names | AMBP, HCP, ITIL | |||
|
Domain Architecture |
|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin (Protein HC) (Complex-forming glycoprotein heterogeneous in charge) (Alpha-1 microglycoprotein); Inter-alpha-trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
VNS2_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 17) | NC score | 0.410553 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62472 | Gene names | Lcn4 | |||
|
Domain Architecture |
|
|||||
| Description | Vomeronasal secretory protein 2 precursor (Vomeronasal secretory protein II) (VNSP II) (Lipocalin-4). | |||||
|
LCN1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 18) | NC score | 0.407980 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P31025 | Gene names | LCN1, VEGP | |||
|
Domain Architecture |
|
|||||
| Description | Lipocalin-1 precursor (Von Ebner gland protein) (VEG protein) (Tear prealbumin) (TP) (Tear lipocalin) (Tlc). | |||||
|
OBP2B_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 19) | NC score | 0.441661 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NPH6, Q9NY51, Q9NY52 | Gene names | OBP2B | |||
|
Domain Architecture |
|
|||||
| Description | Odorant-binding protein 2b precursor (OBPIIb). | |||||
|
NGAL_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 20) | NC score | 0.350275 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P80188, P30150, Q92683 | Gene names | LCN2, HNL, NGAL | |||
|
Domain Architecture |
|
|||||
| Description | Neutrophil gelatinase-associated lipocalin precursor (NGAL) (p25) (25 kDa alpha-2-microglobulin-related subunit of MMP-9) (Lipocalin-2) (Oncogene 24p3). | |||||
|
OBP2A_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 21) | NC score | 0.387800 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NY56, Q9NY50, Q9NY53, Q9NY54, Q9NY55 | Gene names | OBP2A | |||
|
Domain Architecture |
|
|||||
| Description | Odorant-binding protein 2a precursor (OBPIIa). | |||||
|
LCN8_MOUSE
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.149387 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q924P3 | Gene names | Lcn8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-8 precursor (Epididymal 17 kDa lipocalin) (EP17) (mEP17). | |||||
|
LCN12_MOUSE
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.210522 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6JVL5 | Gene names | Lcn12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-12 precursor. | |||||
|
A1AG3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.042332 (rank : 30) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q63805 | Gene names | Orm3, Agp-3, Orm-3 | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-1-acid glycoprotein 3 precursor (AGP 3) (Orosomucoid-3) (OMD 3). | |||||
|
LCN13_MOUSE
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.304500 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K1H9 | Gene names | Lcn13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-13 precursor. | |||||
|
LCN10_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.153362 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q810Z1 | Gene names | Lcn10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-10 precursor. | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.011192 (rank : 31) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
LOX5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.005677 (rank : 32) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48999 | Gene names | Alox5 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase (EC 1.13.11.34) (5-lipoxygenase) (5-LO). | |||||
|
PCDG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.000347 (rank : 33) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5H4, Q9Y5D6 | Gene names | PCDHGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A1 precursor (PCDH-gamma-A1). | |||||
|
CO8G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.082576 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07360, Q5SQ07 | Gene names | C8G | |||
|
Domain Architecture |
|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
CO8G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.111190 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VCG4 | Gene names | C8g | |||
|
Domain Architecture |
|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
LCN12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.126728 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6JVE5 | Gene names | LCN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-12 precursor. | |||||
|
LCN6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.067332 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P62502, Q71SF6 | Gene names | LCN6, LCN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-6 precursor (Lipocalin-5). | |||||
|
MUP5_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.38316e-93 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P11591 | Gene names | Mup5 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 5 precursor (MUP 5). | |||||
|
MUP4_MOUSE
|
||||||
| NC score | 0.996747 (rank : 2) | θ value | 2.55803e-79 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P11590 | Gene names | Mup4 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 4 precursor (MUP 4). | |||||
|
MUP6_MOUSE
|
||||||
| NC score | 0.994550 (rank : 3) | θ value | 2.47765e-74 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P02762, P70119, Q78EF5, Q91V46 | Gene names | Mup6 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 6 precursor (MUP 6) (Alpha-2U-globulin) (Group 1, BS6) (Allergen Mus m 1). | |||||
|
MUP1_MOUSE
|
||||||
| NC score | 0.994533 (rank : 4) | θ value | 7.20884e-74 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11588, Q61921 | Gene names | Mup1 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 1 precursor (MUP 1). | |||||
|
MUP2_MOUSE
|
||||||
| NC score | 0.994344 (rank : 5) | θ value | 4.22625e-74 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11589, Q99JF6 | Gene names | Mup2 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 2 precursor (MUP 2). | |||||
|
MUP3_MOUSE
|
||||||
| NC score | 0.994217 (rank : 6) | θ value | 2.83479e-70 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P04939, P97897, Q8VCG6 | Gene names | Mup3 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 3 precursor (MUP 3) (Non-group 1/group 2 MUP15). | |||||
|
LCN9_HUMAN
|
||||||
| NC score | 0.918141 (rank : 7) | θ value | 5.40856e-29 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WX39, Q6JVE7 | Gene names | LCN9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-9 precursor (MUP-like lipocalin). | |||||
|
LCN9_MOUSE
|
||||||
| NC score | 0.909736 (rank : 8) | θ value | 5.06226e-27 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D267, Q80ZC4 | Gene names | Lcn9 | |||
|
Domain Architecture |

|
|||||
| Description | Epididymal-specific lipocalin-9 precursor (MUP-like lipocalin). | |||||
|
PBAS_MOUSE
|
||||||
| NC score | 0.605885 (rank : 9) | θ value | 1.29631e-06 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08976 | Gene names | Pbsn, Prbs | |||
|
Domain Architecture |

|
|||||
| Description | Probasin precursor (PB). | |||||
|
VNS1_MOUSE
|
||||||
| NC score | 0.601079 (rank : 10) | θ value | 2.13673e-09 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62471 | Gene names | Lcn3 | |||
|
Domain Architecture |

|
|||||
| Description | Vomeronasal secretory protein 1 precursor (Vomeronasal secretory protein I) (VNSP I) (Lipocalin-3). | |||||
|
PAEP_HUMAN
|
||||||
| NC score | 0.568318 (rank : 11) | θ value | 2.44474e-05 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P09466, Q9UG92 | Gene names | PAEP | |||
|
Domain Architecture |

|
|||||
| Description | Glycodelin precursor (GD) (Pregnancy-associated endometrial alpha-2 globulin) (PEG) (PAEG) (Placental protein 14) (PP14) (Progesterone- associated endometrial protein) (Progestagen-associated endometrial protein). | |||||
|
PTGDS_HUMAN
|
||||||
| NC score | 0.554308 (rank : 12) | θ value | 6.87365e-08 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41222, Q5SQ10, Q7M4P3, Q9UCC9, Q9UCD9 | Gene names | PTGDS, PDS | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS) (Beta-trace protein) (Cerebrin-28). | |||||
|
PTGDS_MOUSE
|
||||||
| NC score | 0.525906 (rank : 13) | θ value | 5.81887e-07 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O09114, O09157, O35091, Q62169 | Gene names | Ptgds | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS). | |||||
|
NGAL_MOUSE
|
||||||
| NC score | 0.507710 (rank : 14) | θ value | 7.59969e-07 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P11672 | Gene names | Lcn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil gelatinase-associated lipocalin precursor (NGAL) (p25) (SV- 40-induced 24P3 protein) (Lipocalin-2). | |||||
|
OBP2B_HUMAN
|
||||||
| NC score | 0.441661 (rank : 15) | θ value | 0.00175202 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NPH6, Q9NY51, Q9NY52 | Gene names | OBP2B | |||
|
Domain Architecture |

|
|||||
| Description | Odorant-binding protein 2b precursor (OBPIIb). | |||||
|
VNS2_MOUSE
|
||||||
| NC score | 0.410553 (rank : 16) | θ value | 0.00020696 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62472 | Gene names | Lcn4 | |||
|
Domain Architecture |

|
|||||
| Description | Vomeronasal secretory protein 2 precursor (Vomeronasal secretory protein II) (VNSP II) (Lipocalin-4). | |||||
|
LCN1_HUMAN
|
||||||
| NC score | 0.407980 (rank : 17) | θ value | 0.00134147 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P31025 | Gene names | LCN1, VEGP | |||
|
Domain Architecture |

|
|||||
| Description | Lipocalin-1 precursor (Von Ebner gland protein) (VEG protein) (Tear prealbumin) (TP) (Tear lipocalin) (Tlc). | |||||
|
OBP2A_HUMAN
|
||||||
| NC score | 0.387800 (rank : 18) | θ value | 0.0252991 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NY56, Q9NY50, Q9NY53, Q9NY54, Q9NY55 | Gene names | OBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Odorant-binding protein 2a precursor (OBPIIa). | |||||
|
NGAL_HUMAN
|
||||||
| NC score | 0.350275 (rank : 19) | θ value | 0.00390308 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P80188, P30150, Q92683 | Gene names | LCN2, HNL, NGAL | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil gelatinase-associated lipocalin precursor (NGAL) (p25) (25 kDa alpha-2-microglobulin-related subunit of MMP-9) (Lipocalin-2) (Oncogene 24p3). | |||||
|
AMBP_MOUSE
|
||||||
| NC score | 0.337628 (rank : 20) | θ value | 1.87187e-05 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07456, Q61294 | Gene names | Ambp, Itil | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin; Inter-alpha- trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
AMBP_HUMAN
|
||||||
| NC score | 0.334853 (rank : 21) | θ value | 3.19293e-05 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P02760, P00977, P02759 | Gene names | AMBP, HCP, ITIL | |||
|
Domain Architecture |

|
|||||
| Description | AMBP protein precursor [Contains: Alpha-1-microglobulin (Protein HC) (Complex-forming glycoprotein heterogeneous in charge) (Alpha-1 microglycoprotein); Inter-alpha-trypsin inhibitor light chain (ITI-LC) (Bikunin) (HI-30)]. | |||||
|
LCN13_MOUSE
|
||||||
| NC score | 0.304500 (rank : 22) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K1H9 | Gene names | Lcn13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-13 precursor. | |||||
|
LCN12_MOUSE
|
||||||
| NC score | 0.210522 (rank : 23) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6JVL5 | Gene names | Lcn12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-12 precursor. | |||||
|
LCN10_MOUSE
|
||||||
| NC score | 0.153362 (rank : 24) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q810Z1 | Gene names | Lcn10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-10 precursor. | |||||
|
LCN8_MOUSE
|
||||||
| NC score | 0.149387 (rank : 25) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q924P3 | Gene names | Lcn8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-8 precursor (Epididymal 17 kDa lipocalin) (EP17) (mEP17). | |||||
|
LCN12_HUMAN
|
||||||
| NC score | 0.126728 (rank : 26) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6JVE5 | Gene names | LCN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-12 precursor. | |||||
|
CO8G_MOUSE
|
||||||
| NC score | 0.111190 (rank : 27) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VCG4 | Gene names | C8g | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
CO8G_HUMAN
|
||||||
| NC score | 0.082576 (rank : 28) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07360, Q5SQ07 | Gene names | C8G | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
LCN6_HUMAN
|
||||||
| NC score | 0.067332 (rank : 29) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P62502, Q71SF6 | Gene names | LCN6, LCN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-6 precursor (Lipocalin-5). | |||||
|
A1AG3_MOUSE
|
||||||
| NC score | 0.042332 (rank : 30) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q63805 | Gene names | Orm3, Agp-3, Orm-3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-acid glycoprotein 3 precursor (AGP 3) (Orosomucoid-3) (OMD 3). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.011192 (rank : 31) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
LOX5_MOUSE
|
||||||
| NC score | 0.005677 (rank : 32) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48999 | Gene names | Alox5 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase (EC 1.13.11.34) (5-lipoxygenase) (5-LO). | |||||
|
PCDG1_HUMAN
|
||||||
| NC score | 0.000347 (rank : 33) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5H4, Q9Y5D6 | Gene names | PCDHGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A1 precursor (PCDH-gamma-A1). | |||||