Please be patient as the page loads
|
LCN8_MOUSE
|
||||||
| SwissProt Accessions | Q924P3 | Gene names | Lcn8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-8 precursor (Epididymal 17 kDa lipocalin) (EP17) (mEP17). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LCN8_MOUSE
|
||||||
| θ value | 6.06035e-97 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q924P3 | Gene names | Lcn8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-8 precursor (Epididymal 17 kDa lipocalin) (EP17) (mEP17). | |||||
|
LCN8_HUMAN
|
||||||
| θ value | 1.97229e-55 (rank : 2) | NC score | 0.952566 (rank : 2) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6JVE9, Q5T5R4 | Gene names | LCN8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-8 precursor. | |||||
|
CO8G_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 3) | NC score | 0.138516 (rank : 9) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCG4 | Gene names | C8g | |||
|
Domain Architecture |
|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
PAEP_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 4) | NC score | 0.230221 (rank : 3) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P09466, Q9UG92 | Gene names | PAEP | |||
|
Domain Architecture |
|
|||||
| Description | Glycodelin precursor (GD) (Pregnancy-associated endometrial alpha-2 globulin) (PEG) (PAEG) (Placental protein 14) (PP14) (Progesterone- associated endometrial protein) (Progestagen-associated endometrial protein). | |||||
|
LCN9_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 5) | NC score | 0.221672 (rank : 4) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D267, Q80ZC4 | Gene names | Lcn9 | |||
|
Domain Architecture |
|
|||||
| Description | Epididymal-specific lipocalin-9 precursor (MUP-like lipocalin). | |||||
|
MUP3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 6) | NC score | 0.157683 (rank : 6) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P04939, P97897, Q8VCG6 | Gene names | Mup3 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 3 precursor (MUP 3) (Non-group 1/group 2 MUP15). | |||||
|
MUP4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 7) | NC score | 0.153345 (rank : 7) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11590 | Gene names | Mup4 | |||
|
Domain Architecture |
|
|||||
| Description | Major urinary protein 4 precursor (MUP 4). | |||||
|
MUP5_MOUSE
|
||||||
| θ value | 0.279714 (rank : 8) | NC score | 0.149387 (rank : 8) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11591 | Gene names | Mup5 | |||
|
Domain Architecture |
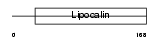
|
|||||
| Description | Major urinary protein 5 precursor (MUP 5). | |||||
|
LCN10_MOUSE
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.124496 (rank : 10) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q810Z1 | Gene names | Lcn10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-10 precursor. | |||||
|
LCN9_HUMAN
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.171369 (rank : 5) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WX39, Q6JVE7 | Gene names | LCN9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-9 precursor (MUP-like lipocalin). | |||||
|
LCN6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 11) | NC score | 0.085181 (rank : 14) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62502, Q71SF6 | Gene names | LCN6, LCN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-6 precursor (Lipocalin-5). | |||||
|
CO8G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 12) | NC score | 0.061460 (rank : 18) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P07360, Q5SQ07 | Gene names | C8G | |||
|
Domain Architecture |
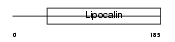
|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
LCN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 13) | NC score | 0.068225 (rank : 17) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P31025 | Gene names | LCN1, VEGP | |||
|
Domain Architecture |
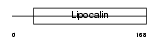
|
|||||
| Description | Lipocalin-1 precursor (Von Ebner gland protein) (VEG protein) (Tear prealbumin) (TP) (Tear lipocalin) (Tlc). | |||||
|
MUP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 14) | NC score | 0.117808 (rank : 11) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11588, Q61921 | Gene names | Mup1 | |||
|
Domain Architecture |
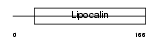
|
|||||
| Description | Major urinary protein 1 precursor (MUP 1). | |||||
|
MUP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 15) | NC score | 0.117059 (rank : 13) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11589, Q99JF6 | Gene names | Mup2 | |||
|
Domain Architecture |
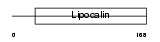
|
|||||
| Description | Major urinary protein 2 precursor (MUP 2). | |||||
|
MUP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 16) | NC score | 0.117727 (rank : 12) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02762, P70119, Q78EF5, Q91V46 | Gene names | Mup6 | |||
|
Domain Architecture |
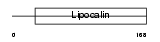
|
|||||
| Description | Major urinary protein 6 precursor (MUP 6) (Alpha-2U-globulin) (Group 1, BS6) (Allergen Mus m 1). | |||||
|
PBAS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 17) | NC score | 0.069196 (rank : 16) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08976 | Gene names | Pbsn, Prbs | |||
|
Domain Architecture |
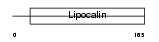
|
|||||
| Description | Probasin precursor (PB). | |||||
|
PTGDS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 18) | NC score | 0.083408 (rank : 15) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41222, Q5SQ10, Q7M4P3, Q9UCC9, Q9UCD9 | Gene names | PTGDS, PDS | |||
|
Domain Architecture |
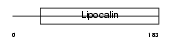
|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS) (Beta-trace protein) (Cerebrin-28). | |||||
|
PTGDS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 19) | NC score | 0.059278 (rank : 20) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O09114, O09157, O35091, Q62169 | Gene names | Ptgds | |||
|
Domain Architecture |
|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS). | |||||
|
VNS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 20) | NC score | 0.059842 (rank : 19) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62471 | Gene names | Lcn3 | |||
|
Domain Architecture |
|
|||||
| Description | Vomeronasal secretory protein 1 precursor (Vomeronasal secretory protein I) (VNSP I) (Lipocalin-3). | |||||
|
LCN8_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.06035e-97 (rank : 1) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q924P3 | Gene names | Lcn8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-8 precursor (Epididymal 17 kDa lipocalin) (EP17) (mEP17). | |||||
|
LCN8_HUMAN
|
||||||
| NC score | 0.952566 (rank : 2) | θ value | 1.97229e-55 (rank : 2) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6JVE9, Q5T5R4 | Gene names | LCN8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-8 precursor. | |||||
|
PAEP_HUMAN
|
||||||
| NC score | 0.230221 (rank : 3) | θ value | 0.0148317 (rank : 4) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P09466, Q9UG92 | Gene names | PAEP | |||
|
Domain Architecture |

|
|||||
| Description | Glycodelin precursor (GD) (Pregnancy-associated endometrial alpha-2 globulin) (PEG) (PAEG) (Placental protein 14) (PP14) (Progesterone- associated endometrial protein) (Progestagen-associated endometrial protein). | |||||
|
LCN9_MOUSE
|
||||||
| NC score | 0.221672 (rank : 4) | θ value | 0.0193708 (rank : 5) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D267, Q80ZC4 | Gene names | Lcn9 | |||
|
Domain Architecture |

|
|||||
| Description | Epididymal-specific lipocalin-9 precursor (MUP-like lipocalin). | |||||
|
LCN9_HUMAN
|
||||||
| NC score | 0.171369 (rank : 5) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WX39, Q6JVE7 | Gene names | LCN9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-9 precursor (MUP-like lipocalin). | |||||
|
MUP3_MOUSE
|
||||||
| NC score | 0.157683 (rank : 6) | θ value | 0.0736092 (rank : 6) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P04939, P97897, Q8VCG6 | Gene names | Mup3 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 3 precursor (MUP 3) (Non-group 1/group 2 MUP15). | |||||
|
MUP4_MOUSE
|
||||||
| NC score | 0.153345 (rank : 7) | θ value | 0.125558 (rank : 7) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11590 | Gene names | Mup4 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 4 precursor (MUP 4). | |||||
|
MUP5_MOUSE
|
||||||
| NC score | 0.149387 (rank : 8) | θ value | 0.279714 (rank : 8) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11591 | Gene names | Mup5 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 5 precursor (MUP 5). | |||||
|
CO8G_MOUSE
|
||||||
| NC score | 0.138516 (rank : 9) | θ value | 0.0113563 (rank : 3) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCG4 | Gene names | C8g | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
LCN10_MOUSE
|
||||||
| NC score | 0.124496 (rank : 10) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q810Z1 | Gene names | Lcn10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-10 precursor. | |||||
|
MUP1_MOUSE
|
||||||
| NC score | 0.117808 (rank : 11) | θ value | θ > 10 (rank : 14) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11588, Q61921 | Gene names | Mup1 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 1 precursor (MUP 1). | |||||
|
MUP6_MOUSE
|
||||||
| NC score | 0.117727 (rank : 12) | θ value | θ > 10 (rank : 16) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02762, P70119, Q78EF5, Q91V46 | Gene names | Mup6 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 6 precursor (MUP 6) (Alpha-2U-globulin) (Group 1, BS6) (Allergen Mus m 1). | |||||
|
MUP2_MOUSE
|
||||||
| NC score | 0.117059 (rank : 13) | θ value | θ > 10 (rank : 15) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11589, Q99JF6 | Gene names | Mup2 | |||
|
Domain Architecture |

|
|||||
| Description | Major urinary protein 2 precursor (MUP 2). | |||||
|
LCN6_HUMAN
|
||||||
| NC score | 0.085181 (rank : 14) | θ value | 1.81305 (rank : 11) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62502, Q71SF6 | Gene names | LCN6, LCN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epididymal-specific lipocalin-6 precursor (Lipocalin-5). | |||||
|
PTGDS_HUMAN
|
||||||
| NC score | 0.083408 (rank : 15) | θ value | θ > 10 (rank : 18) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41222, Q5SQ10, Q7M4P3, Q9UCC9, Q9UCD9 | Gene names | PTGDS, PDS | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS) (Beta-trace protein) (Cerebrin-28). | |||||
|
PBAS_MOUSE
|
||||||
| NC score | 0.069196 (rank : 16) | θ value | θ > 10 (rank : 17) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08976 | Gene names | Pbsn, Prbs | |||
|
Domain Architecture |

|
|||||
| Description | Probasin precursor (PB). | |||||
|
LCN1_HUMAN
|
||||||
| NC score | 0.068225 (rank : 17) | θ value | θ > 10 (rank : 13) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P31025 | Gene names | LCN1, VEGP | |||
|
Domain Architecture |

|
|||||
| Description | Lipocalin-1 precursor (Von Ebner gland protein) (VEG protein) (Tear prealbumin) (TP) (Tear lipocalin) (Tlc). | |||||
|
CO8G_HUMAN
|
||||||
| NC score | 0.061460 (rank : 18) | θ value | θ > 10 (rank : 12) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P07360, Q5SQ07 | Gene names | C8G | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 gamma chain precursor. | |||||
|
VNS1_MOUSE
|
||||||
| NC score | 0.059842 (rank : 19) | θ value | θ > 10 (rank : 20) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62471 | Gene names | Lcn3 | |||
|
Domain Architecture |

|
|||||
| Description | Vomeronasal secretory protein 1 precursor (Vomeronasal secretory protein I) (VNSP I) (Lipocalin-3). | |||||
|
PTGDS_MOUSE
|
||||||
| NC score | 0.059278 (rank : 20) | θ value | θ > 10 (rank : 19) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O09114, O09157, O35091, Q62169 | Gene names | Ptgds | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin-H2 D-isomerase precursor (EC 5.3.99.2) (Lipocalin-type prostaglandin-D synthase) (Glutathione-independent PGD synthetase) (Prostaglandin-D2 synthase) (PGD2 synthase) (PGDS2) (PGDS). | |||||