Please be patient as the page loads
|
UTP20_HUMAN
|
||||||
| SwissProt Accessions | O75691, Q9H3H4 | Gene names | UTP20, DRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein) (Protein Key-1A6) (Novel nucleolar protein 73) (NNP73). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UTP20_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O75691, Q9H3H4 | Gene names | UTP20, DRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein) (Protein Key-1A6) (Novel nucleolar protein 73) (NNP73). | |||||
|
UTP20_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989022 (rank : 2) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5XG71, Q80V22, Q8BXH9, Q8CHL6, Q99K11 | Gene names | Utp20, Drim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein). | |||||
|
CI093_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 3) | NC score | 0.043231 (rank : 7) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 4) | NC score | 0.054739 (rank : 4) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
IF140_HUMAN
|
||||||
| θ value | 0.163984 (rank : 5) | NC score | 0.061623 (rank : 3) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96RY7, O60332 | Gene names | IFT140, KIAA0590, WDTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 140 homolog (WD and tetratricopeptide repeats protein 2). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 6) | NC score | 0.026316 (rank : 16) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 7) | NC score | 0.038319 (rank : 8) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
TRI39_HUMAN
|
||||||
| θ value | 0.62314 (rank : 8) | NC score | 0.018084 (rank : 33) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HCM9, Q76BL3, Q8IYT9, Q96IB6 | Gene names | TRIM39, RNF23, TFP | |||
|
Domain Architecture |
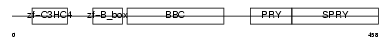
|
|||||
| Description | Tripartite motif-containing protein 39 (RING finger protein 23) (Testis-abundant finger protein). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 9) | NC score | 0.025426 (rank : 19) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
THYG_HUMAN
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.018263 (rank : 32) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
EVPL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.023083 (rank : 21) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
WIPI1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.023983 (rank : 20) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3E3, Q8BGE1, Q8R1A9, Q8R1C7 | Gene names | Wipi1, D11Ertd498e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat domain phosphoinositide-interacting protein 1 (WIPI-1) (WD40 repeat protein interacting with phosphoinositides of 49 kDa) (WIPI 49 kDa). | |||||
|
ABCAD_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.011627 (rank : 55) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
BCAS2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.047795 (rank : 5) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75934, Q6FGS0 | Gene names | BCAS2, DAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 2 (DNA amplified in mammary carcinoma 1 protein) (Spliceosome-associated protein SPF 27). | |||||
|
BCAS2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.047795 (rank : 6) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D287, Q3TKJ5, Q91YX8, Q9D2U4, Q9DAI0 | Gene names | Bcas2, Dam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 2 homolog (DNA amplified in mammary carcinoma 1 protein). | |||||
|
K0494_HUMAN
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.029259 (rank : 13) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75071 | Gene names | KIAA0494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized calcium-binding protein KIAA0494. | |||||
|
NEMO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.031886 (rank : 11) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
PTPRR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.013015 (rank : 52) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62132, Q64491, Q64492, Q9QUH9 | Gene names | Ptprr, Ptp13 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine-phosphatase SL) (Phosphotyrosine phosphatase 13). | |||||
|
CTGE2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.021509 (rank : 23) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96RT6 | Gene names | CTAGE1, CTAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-2. | |||||
|
LRSM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.019425 (rank : 30) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80ZI6 | Gene names | Lrsam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase). | |||||
|
STAT2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.014066 (rank : 43) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
AT8B4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.008866 (rank : 65) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TF62, Q9H727 | Gene names | ATP8B4, KIAA1939 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IM (EC 3.6.3.1) (ATPase class I type 8B member 4). | |||||
|
CI065_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.029909 (rank : 12) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WUY3, Q5T304 | Gene names | C9orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf65. | |||||
|
CNKR2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.026255 (rank : 18) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.026256 (rank : 17) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
RBP17_HUMAN
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.028193 (rank : 14) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H2T7 | Gene names | RANBP17 | |||
|
Domain Architecture |
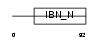
|
|||||
| Description | Ran-binding protein 17. | |||||
|
CAND2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.018792 (rank : 31) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.015408 (rank : 40) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.016745 (rank : 38) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.017744 (rank : 34) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
NF2L3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.021063 (rank : 25) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
NF2L3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.021709 (rank : 22) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
ZPBP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.020753 (rank : 27) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BS86, Q15941, Q75KX9, Q75MI3 | Gene names | ZPBP, ZPBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zona pellucida-binding protein 1 precursor (Sp38). | |||||
|
BRD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.009431 (rank : 63) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BTAF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.013196 (rank : 51) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
DHX9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.010735 (rank : 58) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
ETV4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.008817 (rank : 66) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
EXOC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.017562 (rank : 35) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96KP1, Q5JPC8, Q96AN6, Q9NUZ8, Q9UJM7 | Gene names | EXOC2, SEC5, SEC5L1 | |||
|
Domain Architecture |
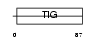
|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.013314 (rank : 49) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
PRI1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.020078 (rank : 28) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49642 | Gene names | PRIM1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA primase small subunit (EC 2.7.7.-) (DNA primase 49 kDa subunit) (p49). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.008080 (rank : 67) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
ANKR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.013227 (rank : 50) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CR42, O55014, Q3UIF7, Q3UJ39, Q792Q9 | Gene names | Ankrd1, Carp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.013631 (rank : 45) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
OS9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.035550 (rank : 9) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13438, O00579 | Gene names | OS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OS-9 precursor (Amplified in osteosarcoma 9). | |||||
|
PDE1C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.006186 (rank : 68) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
POPD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.033456 (rank : 10) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ES83, Q8C8L3 | Gene names | Bves, Pop1, Popdc1 | |||
|
Domain Architecture |
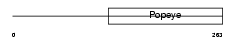
|
|||||
| Description | Blood vessel epicardial substance (Popeye domain-containing protein 1) (Popeye protein 1). | |||||
|
SENP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.011706 (rank : 54) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59110 | Gene names | Senp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
ALG12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.013404 (rank : 46) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BV10, Q8NG10, Q96AA4 | Gene names | ALG12 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichyl-P-Man:Man(7)GlcNAc(2)-PP-dolichyl-alpha-1,6- mannosyltransferase (EC 2.4.1.-) (Mannosyltransferase ALG12 homolog) (hALG12) (Membrane protein SB87). | |||||
|
ANKR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.010760 (rank : 57) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15327, Q96LE7 | Gene names | ANKRD1, C193, CARP, HA1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein) (Cytokine-inducible nuclear protein) (C-193). | |||||
|
ATR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.019551 (rank : 29) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
CAND2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.015512 (rank : 39) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75155 | Gene names | CAND2, KIAA0667 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2). | |||||
|
CLPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.021222 (rank : 24) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16740 | Gene names | CLPP | |||
|
Domain Architecture |
|
|||||
| Description | Putative ATP-dependent Clp protease proteolytic subunit, mitochondrial precursor (EC 3.4.21.92) (Endopeptidase Clp). | |||||
|
CLPP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.020978 (rank : 26) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88696, Q3TI13 | Gene names | Clpp | |||
|
Domain Architecture |
|
|||||
| Description | Putative ATP-dependent Clp protease proteolytic subunit, mitochondrial precursor (EC 3.4.21.92) (Endopeptidase Clp). | |||||
|
CPSF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.011586 (rank : 56) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKF6, O14769, Q96F36 | Gene names | CPSF3, CPSF73 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 73 kDa subunit (CPSF 73 kDa subunit). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.015179 (rank : 41) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
DHX9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.009274 (rank : 64) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.014002 (rank : 44) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
HEAT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.017319 (rank : 36) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86Y56, Q69YL1, Q96FI9, Q9NX75 | Gene names | HEATR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 2. | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.010437 (rank : 59) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
MBD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.012664 (rank : 53) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2D7, Q792D2, Q8R3R3 | Gene names | Mbd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 4 (EC 3.2.2.-) (Methyl-CpG-binding protein MBD4) (Mismatch-specific DNA N-glycosylase). | |||||
|
NEBU_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.009765 (rank : 62) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20929, Q15346 | Gene names | NEB | |||
|
Domain Architecture |
|
|||||
| Description | Nebulin. | |||||
|
NOC2L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.014247 (rank : 42) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WV70, Q8VE16 | Gene names | Noc2l | |||
|
Domain Architecture |
|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
PKN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.001488 (rank : 69) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.010056 (rank : 61) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
RBP17_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.026907 (rank : 15) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99NF8, Q9D4E3 | Gene names | Ranbp17 | |||
|
Domain Architecture |
|
|||||
| Description | Ran-binding protein 17. | |||||
|
STMN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.013361 (rank : 47) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q93045, O14952, Q6PK68 | Gene names | STMN2, SCG10, SCGN10 | |||
|
Domain Architecture |
|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
STMN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.013361 (rank : 48) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P55821 | Gene names | Stmn2, Scg10, Scgn10, Stmb2 | |||
|
Domain Architecture |
|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
TRAIP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.017001 (rank : 37) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
TXND3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.010372 (rank : 60) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N427, Q9NZH1 | Gene names | TXNDC3, NME8, SPTRX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 3 (Spermatid-specific thioredoxin-2) (Sptrx-2) (NM23-H8). | |||||
|
UTP20_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O75691, Q9H3H4 | Gene names | UTP20, DRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein) (Protein Key-1A6) (Novel nucleolar protein 73) (NNP73). | |||||
|
UTP20_MOUSE
|
||||||
| NC score | 0.989022 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5XG71, Q80V22, Q8BXH9, Q8CHL6, Q99K11 | Gene names | Utp20, Drim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein). | |||||
|
IF140_HUMAN
|
||||||
| NC score | 0.061623 (rank : 3) | θ value | 0.163984 (rank : 5) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96RY7, O60332 | Gene names | IFT140, KIAA0590, WDTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 140 homolog (WD and tetratricopeptide repeats protein 2). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.054739 (rank : 4) | θ value | 0.0252991 (rank : 4) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
BCAS2_HUMAN
|
||||||
| NC score | 0.047795 (rank : 5) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75934, Q6FGS0 | Gene names | BCAS2, DAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 2 (DNA amplified in mammary carcinoma 1 protein) (Spliceosome-associated protein SPF 27). | |||||
|
BCAS2_MOUSE
|
||||||
| NC score | 0.047795 (rank : 6) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D287, Q3TKJ5, Q91YX8, Q9D2U4, Q9DAI0 | Gene names | Bcas2, Dam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 2 homolog (DNA amplified in mammary carcinoma 1 protein). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.043231 (rank : 7) | θ value | 0.0148317 (rank : 3) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.038319 (rank : 8) | θ value | 0.62314 (rank : 7) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
OS9_HUMAN
|
||||||
| NC score | 0.035550 (rank : 9) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13438, O00579 | Gene names | OS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OS-9 precursor (Amplified in osteosarcoma 9). | |||||
|
POPD1_MOUSE
|
||||||
| NC score | 0.033456 (rank : 10) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ES83, Q8C8L3 | Gene names | Bves, Pop1, Popdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Blood vessel epicardial substance (Popeye domain-containing protein 1) (Popeye protein 1). | |||||
|
NEMO_HUMAN
|
||||||
| NC score | 0.031886 (rank : 11) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
CI065_HUMAN
|
||||||
| NC score | 0.029909 (rank : 12) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WUY3, Q5T304 | Gene names | C9orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf65. | |||||
|
K0494_HUMAN
|
||||||
| NC score | 0.029259 (rank : 13) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75071 | Gene names | KIAA0494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized calcium-binding protein KIAA0494. | |||||
|
RBP17_HUMAN
|
||||||
| NC score | 0.028193 (rank : 14) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H2T7 | Gene names | RANBP17 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 17. | |||||
|
RBP17_MOUSE
|
||||||
| NC score | 0.026907 (rank : 15) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99NF8, Q9D4E3 | Gene names | Ranbp17 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 17. | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.026316 (rank : 16) | θ value | 0.47712 (rank : 6) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
CNKR2_MOUSE
|
||||||
| NC score | 0.026256 (rank : 17) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_HUMAN
|
||||||
| NC score | 0.026255 (rank : 18) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.025426 (rank : 19) | θ value | 1.06291 (rank : 9) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
WIPI1_MOUSE
|
||||||
| NC score | 0.023983 (rank : 20) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3E3, Q8BGE1, Q8R1A9, Q8R1C7 | Gene names | Wipi1, D11Ertd498e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat domain phosphoinositide-interacting protein 1 (WIPI-1) (WD40 repeat protein interacting with phosphoinositides of 49 kDa) (WIPI 49 kDa). | |||||
|
EVPL_MOUSE
|
||||||
| NC score | 0.023083 (rank : 21) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
NF2L3_MOUSE
|
||||||
| NC score | 0.021709 (rank : 22) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
CTGE2_HUMAN
|
||||||
| NC score | 0.021509 (rank : 23) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96RT6 | Gene names | CTAGE1, CTAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-2. | |||||
|
CLPP_HUMAN
|
||||||
| NC score | 0.021222 (rank : 24) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16740 | Gene names | CLPP | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent Clp protease proteolytic subunit, mitochondrial precursor (EC 3.4.21.92) (Endopeptidase Clp). | |||||
|
NF2L3_HUMAN
|
||||||
| NC score | 0.021063 (rank : 25) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
CLPP_MOUSE
|
||||||
| NC score | 0.020978 (rank : 26) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88696, Q3TI13 | Gene names | Clpp | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent Clp protease proteolytic subunit, mitochondrial precursor (EC 3.4.21.92) (Endopeptidase Clp). | |||||
|
ZPBP1_HUMAN
|
||||||
| NC score | 0.020753 (rank : 27) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BS86, Q15941, Q75KX9, Q75MI3 | Gene names | ZPBP, ZPBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zona pellucida-binding protein 1 precursor (Sp38). | |||||
|
PRI1_HUMAN
|
||||||
| NC score | 0.020078 (rank : 28) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49642 | Gene names | PRIM1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA primase small subunit (EC 2.7.7.-) (DNA primase 49 kDa subunit) (p49). | |||||
|
ATR_MOUSE
|
||||||
| NC score | 0.019551 (rank : 29) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
LRSM1_MOUSE
|
||||||
| NC score | 0.019425 (rank : 30) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80ZI6 | Gene names | Lrsam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase). | |||||
|
CAND2_MOUSE
|
||||||
| NC score | 0.018792 (rank : 31) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
THYG_HUMAN
|
||||||
| NC score | 0.018263 (rank : 32) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
TRI39_HUMAN
|
||||||
| NC score | 0.018084 (rank : 33) | θ value | 0.62314 (rank : 8) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HCM9, Q76BL3, Q8IYT9, Q96IB6 | Gene names | TRIM39, RNF23, TFP | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 39 (RING finger protein 23) (Testis-abundant finger protein). | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.017744 (rank : 34) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
EXOC2_HUMAN
|
||||||
| NC score | 0.017562 (rank : 35) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96KP1, Q5JPC8, Q96AN6, Q9NUZ8, Q9UJM7 | Gene names | EXOC2, SEC5, SEC5L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
HEAT2_HUMAN
|
||||||
| NC score | 0.017319 (rank : 36) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86Y56, Q69YL1, Q96FI9, Q9NX75 | Gene names | HEATR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 2. | |||||
|
TRAIP_MOUSE
|
||||||
| NC score | 0.017001 (rank : 37) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.016745 (rank : 38) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
CAND2_HUMAN
|
||||||
| NC score | 0.015512 (rank : 39) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75155 | Gene names | CAND2, KIAA0667 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.015408 (rank : 40) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.015179 (rank : 41) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
NOC2L_MOUSE
|
||||||
| NC score | 0.014247 (rank : 42) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WV70, Q8VE16 | Gene names | Noc2l | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
STAT2_MOUSE
|
||||||
| NC score | 0.014066 (rank : 43) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.014002 (rank : 44) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.013631 (rank : 45) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
ALG12_HUMAN
|
||||||
| NC score | 0.013404 (rank : 46) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BV10, Q8NG10, Q96AA4 | Gene names | ALG12 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichyl-P-Man:Man(7)GlcNAc(2)-PP-dolichyl-alpha-1,6- mannosyltransferase (EC 2.4.1.-) (Mannosyltransferase ALG12 homolog) (hALG12) (Membrane protein SB87). | |||||
|
STMN2_HUMAN
|
||||||
| NC score | 0.013361 (rank : 47) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q93045, O14952, Q6PK68 | Gene names | STMN2, SCG10, SCGN10 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
STMN2_MOUSE
|
||||||
| NC score | 0.013361 (rank : 48) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P55821 | Gene names | Stmn2, Scg10, Scgn10, Stmb2 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-2 (SCG10 protein) (Superior cervical ganglion-10 protein). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.013314 (rank : 49) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
ANKR1_MOUSE
|
||||||
| NC score | 0.013227 (rank : 50) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CR42, O55014, Q3UIF7, Q3UJ39, Q792Q9 | Gene names | Ankrd1, Carp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein). | |||||
|
BTAF1_HUMAN
|
||||||
| NC score | 0.013196 (rank : 51) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
PTPRR_MOUSE
|
||||||
| NC score | 0.013015 (rank : 52) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62132, Q64491, Q64492, Q9QUH9 | Gene names | Ptprr, Ptp13 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine-phosphatase SL) (Phosphotyrosine phosphatase 13). | |||||
|
MBD4_MOUSE
|
||||||
| NC score | 0.012664 (rank : 53) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2D7, Q792D2, Q8R3R3 | Gene names | Mbd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 4 (EC 3.2.2.-) (Methyl-CpG-binding protein MBD4) (Mismatch-specific DNA N-glycosylase). | |||||
|
SENP1_MOUSE
|
||||||
| NC score | 0.011706 (rank : 54) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59110 | Gene names | Senp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
ABCAD_HUMAN
|
||||||
| NC score | 0.011627 (rank : 55) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
CPSF3_HUMAN
|
||||||
| NC score | 0.011586 (rank : 56) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKF6, O14769, Q96F36 | Gene names | CPSF3, CPSF73 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 73 kDa subunit (CPSF 73 kDa subunit). | |||||
|
ANKR1_HUMAN
|
||||||
| NC score | 0.010760 (rank : 57) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15327, Q96LE7 | Gene names | ANKRD1, C193, CARP, HA1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein) (Cytokine-inducible nuclear protein) (C-193). | |||||
|
DHX9_HUMAN
|
||||||
| NC score | 0.010735 (rank : 58) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.010437 (rank : 59) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
TXND3_HUMAN
|
||||||
| NC score | 0.010372 (rank : 60) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N427, Q9NZH1 | Gene names | TXNDC3, NME8, SPTRX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 3 (Spermatid-specific thioredoxin-2) (Sptrx-2) (NM23-H8). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.010056 (rank : 61) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
NEBU_HUMAN
|
||||||
| NC score | 0.009765 (rank : 62) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20929, Q15346 | Gene names | NEB | |||
|
Domain Architecture |

|
|||||
| Description | Nebulin. | |||||
|
BRD1_HUMAN
|
||||||
| NC score | 0.009431 (rank : 63) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
DHX9_MOUSE
|
||||||
| NC score | 0.009274 (rank : 64) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
AT8B4_HUMAN
|
||||||
| NC score | 0.008866 (rank : 65) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TF62, Q9H727 | Gene names | ATP8B4, KIAA1939 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IM (EC 3.6.3.1) (ATPase class I type 8B member 4). | |||||
|
ETV4_MOUSE
|
||||||
| NC score | 0.008817 (rank : 66) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.008080 (rank : 67) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
PDE1C_HUMAN
|
||||||
| NC score | 0.006186 (rank : 68) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PKN3_MOUSE
|
||||||
| NC score | 0.001488 (rank : 69) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||