Please be patient as the page loads
|
ETV4_MOUSE
|
||||||
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ETV4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993690 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETV1_MOUSE
|
||||||
| θ value | 1.4755e-135 (rank : 3) | NC score | 0.976842 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_HUMAN
|
||||||
| θ value | 4.29305e-135 (rank : 4) | NC score | 0.976116 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV5_HUMAN
|
||||||
| θ value | 4.03687e-117 (rank : 5) | NC score | 0.966686 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 4.03687e-117 (rank : 6) | NC score | 0.945361 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ETS1_HUMAN
|
||||||
| θ value | 1.2049e-28 (rank : 7) | NC score | 0.869952 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| θ value | 1.2049e-28 (rank : 8) | NC score | 0.869669 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ELK4_MOUSE
|
||||||
| θ value | 2.68423e-28 (rank : 9) | NC score | 0.875934 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ETS2_HUMAN
|
||||||
| θ value | 7.80994e-28 (rank : 10) | NC score | 0.867318 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS2_MOUSE
|
||||||
| θ value | 7.80994e-28 (rank : 11) | NC score | 0.866986 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ELK4_HUMAN
|
||||||
| θ value | 1.73987e-27 (rank : 12) | NC score | 0.860734 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELK1_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 13) | NC score | 0.875152 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_MOUSE
|
||||||
| θ value | 4.28545e-26 (rank : 14) | NC score | 0.873109 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK3_HUMAN
|
||||||
| θ value | 1.62847e-25 (rank : 15) | NC score | 0.880055 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELK3_MOUSE
|
||||||
| θ value | 4.73814e-25 (rank : 16) | NC score | 0.877248 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ERG_MOUSE
|
||||||
| θ value | 6.18819e-25 (rank : 17) | NC score | 0.843397 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
FLI1_HUMAN
|
||||||
| θ value | 6.18819e-25 (rank : 18) | NC score | 0.843577 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
FLI1_MOUSE
|
||||||
| θ value | 6.18819e-25 (rank : 19) | NC score | 0.845794 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
ERG_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 20) | NC score | 0.843414 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
ETV2_HUMAN
|
||||||
| θ value | 5.23862e-24 (rank : 21) | NC score | 0.875840 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETV2_MOUSE
|
||||||
| θ value | 3.39556e-23 (rank : 22) | NC score | 0.870772 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
GABPA_MOUSE
|
||||||
| θ value | 3.39556e-23 (rank : 23) | NC score | 0.850003 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
GABPA_HUMAN
|
||||||
| θ value | 7.56453e-23 (rank : 24) | NC score | 0.849000 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
ERF_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 25) | NC score | 0.818438 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| θ value | 1.02238e-19 (rank : 26) | NC score | 0.818106 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 27) | NC score | 0.785141 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV3_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 28) | NC score | 0.808954 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ELF4_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 29) | NC score | 0.787713 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 4.29542e-18 (rank : 30) | NC score | 0.781826 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF1_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 31) | NC score | 0.787370 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF4_MOUSE
|
||||||
| θ value | 9.56915e-18 (rank : 32) | NC score | 0.768211 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
SPDEF_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 33) | NC score | 0.817739 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ELF2_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 34) | NC score | 0.787685 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 35) | NC score | 0.787128 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
SPDEF_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 36) | NC score | 0.815582 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ETV6_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 37) | NC score | 0.780334 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 38) | NC score | 0.781024 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV7_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 39) | NC score | 0.802621 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ELF5_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 40) | NC score | 0.754488 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF5_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 41) | NC score | 0.754237 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 42) | NC score | 0.062896 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 43) | NC score | 0.026472 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
DPOLM_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 44) | NC score | 0.034852 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIW4, Q9JJW9 | Gene names | Polm | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase mu (EC 2.7.7.7) (Pol Mu). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 45) | NC score | 0.024870 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
PDC6I_HUMAN
|
||||||
| θ value | 0.21417 (rank : 46) | NC score | 0.033900 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
SPRR3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.044114 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
THSD1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.023389 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JM61, Q8C7Q9 | Gene names | Thsd1, Tmtsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
PROX1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.026163 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48437, O88478, Q3UPV1, Q543D8 | Gene names | Prox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
ZHX3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.012020 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0Q2, Q80U14 | Gene names | Zhx3, Kiaa0395, Tix1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and homeodomain protein 3 (Triple homeobox protein 1). | |||||
|
IP3KB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.015807 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27987, Q5VWL9, Q5VWM0, Q96BZ2, Q96JS1, Q9UH47 | Gene names | ITPKB | |||
|
Domain Architecture |

|
|||||
| Description | Inositol-trisphosphate 3-kinase B (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase B) (IP3K B) (IP3 3-kinase B) (IP3K-B). | |||||
|
VAX2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.008969 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTP9 | Gene names | Vax2, Vex | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ventral anterior homeobox 2 (Ventral retina homeodomain protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.017582 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.006763 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
LNK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.009521 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UQQ2, O95184 | Gene names | LNK | |||
|
Domain Architecture |
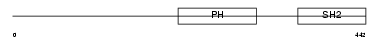
|
|||||
| Description | Lymphocyte-specific adapter protein Lnk (Signal transduction protein Lnk) (Lymphocyte adapter protein). | |||||
|
MBD2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.010541 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2E1, Q811D9, Q9Z2D9 | Gene names | Mbd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 2 (Methyl-CpG-binding protein MBD2). | |||||
|
RIN3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.015978 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
SOX9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.007456 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
DGKK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.015335 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.008185 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.031933 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.033081 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PML_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.010132 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29590, P29591, P29592, P29593, Q00755 | Gene names | PML, MYL, RNF71, TRIM19 | |||
|
Domain Architecture |

|
|||||
| Description | Probable transcription factor PML (Tripartite motif-containing protein 19) (RING finger protein 71). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.014368 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
THSD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.018603 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
TSH2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.005849 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.019644 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.003707 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
FOXJ3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.011577 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.003125 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
NRG2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.004582 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14511 | Gene names | NRG2, NTAK | |||
|
Domain Architecture |

|
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural- and thymus-derived activator for ERBB kinases) (NTAK) (Divergent of neuregulin-1) (DON-1)]. | |||||
|
PROX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.021394 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92786, Q5SW76, Q8TB91 | Gene names | PROX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
SNTB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.007709 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61235 | Gene names | Sntb2, Snt2b2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-2-syntrophin (59 kDa dystrophin-associated protein A1 basic component 2) (Syntrophin 3) (SNT3) (Syntrophin-like) (SNTL). | |||||
|
VAX2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.004796 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UIW0 | Gene names | VAX2 | |||
|
Domain Architecture |
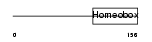
|
|||||
| Description | Ventral anterior homeobox 2. | |||||
|
AATK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | -0.002794 (rank : 104) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80YE4, O35211, Q3U2U5, Q3UHR8, Q66JN3, Q80YE3, Q8CB63, Q8CHE2 | Gene names | Aatk, Kiaa0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase). | |||||
|
ACD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.020546 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96AP0, Q562H5, Q9H8F9 | Gene names | ACD, PIP1, PTOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adrenocortical dysplasia protein homolog (POT1 and TIN2-interacting protein). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.020968 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.025838 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
OS9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.011875 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13438, O00579 | Gene names | OS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OS-9 precursor (Amplified in osteosarcoma 9). | |||||
|
TCF7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.007530 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
UTP20_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.008817 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75691, Q9H3H4 | Gene names | UTP20, DRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein) (Protein Key-1A6) (Novel nucleolar protein 73) (NNP73). | |||||
|
AMHR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | -0.001423 (rank : 103) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16671, Q13762 | Gene names | AMHR2, AMHR | |||
|
Domain Architecture |

|
|||||
| Description | Anti-Muellerian hormone type-2 receptor precursor (EC 2.7.11.30) (Anti-Muellerian hormone type II receptor) (AMH type II receptor) (MIS type II receptor) (MISRII) (MRII). | |||||
|
CNOT4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.012576 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | -0.003270 (rank : 105) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
SIM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.003184 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SPR1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.027185 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.009707 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TJAP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.006032 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5JTD0, Q5JTD1, Q5JWW1, Q68DB2, Q6P2P3, Q9H7V7 | Gene names | TJAP1, PILT, TJP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
CEBPA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.005356 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.013444 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.001178 (rank : 102) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
GSLG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.011909 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61543, Q9QZ40 | Gene names | Glg1, Esl1, Mg160, Selel | |||
|
Domain Architecture |
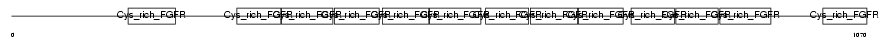
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Selel). | |||||
|
MILK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.005464 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
REST_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | -0.004378 (rank : 106) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
SOX9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.006783 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.009727 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
TAB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.012119 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
TAB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.010767 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
VTNC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.003662 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.001563 (rank : 101) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
SPI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.492720 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.493602 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPIB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.495908 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.480797 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.413512 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPIC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.387943 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
ETV4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETV4_HUMAN
|
||||||
| NC score | 0.993690 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV1_MOUSE
|
||||||
| NC score | 0.976842 (rank : 3) | θ value | 1.4755e-135 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_HUMAN
|
||||||
| NC score | 0.976116 (rank : 4) | θ value | 4.29305e-135 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.966686 (rank : 5) | θ value | 4.03687e-117 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.945361 (rank : 6) | θ value | 4.03687e-117 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ELK3_HUMAN
|
||||||
| NC score | 0.880055 (rank : 7) | θ value | 1.62847e-25 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELK3_MOUSE
|
||||||
| NC score | 0.877248 (rank : 8) | θ value | 4.73814e-25 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ELK4_MOUSE
|
||||||
| NC score | 0.875934 (rank : 9) | θ value | 2.68423e-28 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ETV2_HUMAN
|
||||||
| NC score | 0.875840 (rank : 10) | θ value | 5.23862e-24 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ELK1_HUMAN
|
||||||
| NC score | 0.875152 (rank : 11) | θ value | 4.28545e-26 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_MOUSE
|
||||||
| NC score | 0.873109 (rank : 12) | θ value | 4.28545e-26 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ETV2_MOUSE
|
||||||
| NC score | 0.870772 (rank : 13) | θ value | 3.39556e-23 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETS1_HUMAN
|
||||||
| NC score | 0.869952 (rank : 14) | θ value | 1.2049e-28 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| NC score | 0.869669 (rank : 15) | θ value | 1.2049e-28 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS2_HUMAN
|
||||||
| NC score | 0.867318 (rank : 16) | θ value | 7.80994e-28 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS2_MOUSE
|
||||||
| NC score | 0.866986 (rank : 17) | θ value | 7.80994e-28 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.860734 (rank : 18) | θ value | 1.73987e-27 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
GABPA_MOUSE
|
||||||
| NC score | 0.850003 (rank : 19) | θ value | 3.39556e-23 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
GABPA_HUMAN
|
||||||
| NC score | 0.849000 (rank : 20) | θ value | 7.56453e-23 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
FLI1_MOUSE
|
||||||
| NC score | 0.845794 (rank : 21) | θ value | 6.18819e-25 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
FLI1_HUMAN
|
||||||
| NC score | 0.843577 (rank : 22) | θ value | 6.18819e-25 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
ERG_HUMAN
|
||||||
| NC score | 0.843414 (rank : 23) | θ value | 1.05554e-24 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
ERG_MOUSE
|
||||||
| NC score | 0.843397 (rank : 24) | θ value | 6.18819e-25 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ERF_HUMAN
|
||||||
| NC score | 0.818438 (rank : 25) | θ value | 1.02238e-19 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| NC score | 0.818106 (rank : 26) | θ value | 1.02238e-19 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
SPDEF_HUMAN
|
||||||
| NC score | 0.817739 (rank : 27) | θ value | 1.38178e-16 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
SPDEF_MOUSE
|
||||||
| NC score | 0.815582 (rank : 28) | θ value | 1.80466e-16 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ETV3_MOUSE
|
||||||
| NC score | 0.808954 (rank : 29) | θ value | 3.88503e-19 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV7_HUMAN
|
||||||
| NC score | 0.802621 (rank : 30) | θ value | 4.59992e-12 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ELF4_HUMAN
|
||||||
| NC score | 0.787713 (rank : 31) | θ value | 3.28887e-18 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF2_HUMAN
|
||||||
| NC score | 0.787685 (rank : 32) | θ value | 1.80466e-16 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.787370 (rank : 33) | θ value | 5.60996e-18 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF2_MOUSE
|
||||||
| NC score | 0.787128 (rank : 34) | θ value | 1.80466e-16 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.785141 (rank : 35) | θ value | 3.88503e-19 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.781826 (rank : 36) | θ value | 4.29542e-18 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ETV6_MOUSE
|
||||||
| NC score | 0.781024 (rank : 37) | θ value | 1.68911e-14 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_HUMAN
|
||||||
| NC score | 0.780334 (rank : 38) | θ value | 4.44505e-15 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ELF4_MOUSE
|
||||||
| NC score | 0.768211 (rank : 39) | θ value | 9.56915e-18 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF5_HUMAN
|
||||||
| NC score | 0.754488 (rank : 40) | θ value | 1.133e-10 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF5_MOUSE
|
||||||
| NC score | 0.754237 (rank : 41) | θ value | 1.133e-10 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
SPIB_HUMAN
|
||||||
| NC score | 0.495908 (rank : 42) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPI1_MOUSE
|
||||||
| NC score | 0.493602 (rank : 43) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPI1_HUMAN
|
||||||
| NC score | 0.492720 (rank : 44) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPIB_MOUSE
|
||||||
| NC score | 0.480797 (rank : 45) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIC_HUMAN
|
||||||
| NC score | 0.413512 (rank : 46) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPIC_MOUSE
|
||||||
| NC score | 0.387943 (rank : 47) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.062896 (rank : 48) | θ value | 0.0193708 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
SPRR3_MOUSE
|
||||||
| NC score | 0.044114 (rank : 49) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
DPOLM_MOUSE
|
||||||
| NC score | 0.034852 (rank : 50) | θ value | 0.0736092 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIW4, Q9JJW9 | Gene names | Polm | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase mu (EC 2.7.7.7) (Pol Mu). | |||||
|
PDC6I_HUMAN
|
||||||
| NC score | 0.033900 (rank : 51) | θ value | 0.21417 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.033081 (rank : 52) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.031933 (rank : 53) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SPR1A_HUMAN
|
||||||
| NC score | 0.027185 (rank : 54) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.026472 (rank : 55) | θ value | 0.0563607 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
PROX1_MOUSE
|
||||||
| NC score | 0.026163 (rank : 56) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48437, O88478, Q3UPV1, Q543D8 | Gene names | Prox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.025838 (rank : 57) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.024870 (rank : 58) | θ value | 0.163984 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
THSD1_MOUSE
|
||||||
| NC score | 0.023389 (rank : 59) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JM61, Q8C7Q9 | Gene names | Thsd1, Tmtsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
PROX1_HUMAN
|
||||||
| NC score | 0.021394 (rank : 60) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92786, Q5SW76, Q8TB91 | Gene names | PROX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.020968 (rank : 61) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
ACD_HUMAN
|
||||||
| NC score | 0.020546 (rank : 62) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96AP0, Q562H5, Q9H8F9 | Gene names | ACD, PIP1, PTOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adrenocortical dysplasia protein homolog (POT1 and TIN2-interacting protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.019644 (rank : 63) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
THSD1_HUMAN
|
||||||
| NC score | 0.018603 (rank : 64) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.017582 (rank : 65) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RIN3_HUMAN
|
||||||
| NC score | 0.015978 (rank : 66) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
IP3KB_HUMAN
|
||||||
| NC score | 0.015807 (rank : 67) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27987, Q5VWL9, Q5VWM0, Q96BZ2, Q96JS1, Q9UH47 | Gene names | ITPKB | |||
|
Domain Architecture |

|
|||||
| Description | Inositol-trisphosphate 3-kinase B (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase B) (IP3K B) (IP3 3-kinase B) (IP3K-B). | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.015335 (rank : 68) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.014368 (rank : 69) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.013444 (rank : 70) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CNOT4_HUMAN
|
||||||
| NC score | 0.012576 (rank : 71) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
TAB3_HUMAN
|
||||||
| NC score | 0.012119 (rank : 72) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
ZHX3_MOUSE
|
||||||
| NC score | 0.012020 (rank : 73) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0Q2, Q80U14 | Gene names | Zhx3, Kiaa0395, Tix1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and homeodomain protein 3 (Triple homeobox protein 1). | |||||
|
GSLG1_MOUSE
|
||||||
| NC score | 0.011909 (rank : 74) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61543, Q9QZ40 | Gene names | Glg1, Esl1, Mg160, Selel | |||
|
Domain Architecture |
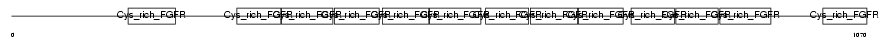
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Selel). | |||||
|
OS9_HUMAN
|
||||||
| NC score | 0.011875 (rank : 75) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13438, O00579 | Gene names | OS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OS-9 precursor (Amplified in osteosarcoma 9). | |||||
|
FOXJ3_HUMAN
|
||||||
| NC score | 0.011577 (rank : 76) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
TAB3_MOUSE
|
||||||
| NC score | 0.010767 (rank : 77) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
MBD2_MOUSE
|
||||||
| NC score | 0.010541 (rank : 78) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2E1, Q811D9, Q9Z2D9 | Gene names | Mbd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 2 (Methyl-CpG-binding protein MBD2). | |||||
|
PML_HUMAN
|
||||||
| NC score | 0.010132 (rank : 79) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29590, P29591, P29592, P29593, Q00755 | Gene names | PML, MYL, RNF71, TRIM19 | |||
|
Domain Architecture |

|
|||||
| Description | Probable transcription factor PML (Tripartite motif-containing protein 19) (RING finger protein 71). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.009727 (rank : 80) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.009707 (rank : 81) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
LNK_HUMAN
|
||||||
| NC score | 0.009521 (rank : 82) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UQQ2, O95184 | Gene names | LNK | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte-specific adapter protein Lnk (Signal transduction protein Lnk) (Lymphocyte adapter protein). | |||||
|
VAX2_MOUSE
|
||||||
| NC score | 0.008969 (rank : 83) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTP9 | Gene names | Vax2, Vex | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ventral anterior homeobox 2 (Ventral retina homeodomain protein). | |||||
|
UTP20_HUMAN
|
||||||
| NC score | 0.008817 (rank : 84) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75691, Q9H3H4 | Gene names | UTP20, DRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein) (Protein Key-1A6) (Novel nucleolar protein 73) (NNP73). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.008185 (rank : 85) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SNTB2_MOUSE
|
||||||
| NC score | 0.007709 (rank : 86) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61235 | Gene names | Sntb2, Snt2b2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-2-syntrophin (59 kDa dystrophin-associated protein A1 basic component 2) (Syntrophin 3) (SNT3) (Syntrophin-like) (SNTL). | |||||
|
TCF7_HUMAN
|
||||||
| NC score | 0.007530 (rank : 87) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
SOX9_HUMAN
|
||||||
| NC score | 0.007456 (rank : 88) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX9_MOUSE
|
||||||
| NC score | 0.006783 (rank : 89) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.006763 (rank : 90) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
TJAP1_HUMAN
|
||||||
| NC score | 0.006032 (rank : 91) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5JTD0, Q5JTD1, Q5JWW1, Q68DB2, Q6P2P3, Q9H7V7 | Gene names | TJAP1, PILT, TJP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
TSH2_MOUSE
|
||||||
| NC score | 0.005849 (rank : 92) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
MILK1_HUMAN
|
||||||
| NC score | 0.005464 (rank : 93) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
CEBPA_MOUSE
|
||||||
| NC score | 0.005356 (rank : 94) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
VAX2_HUMAN
|
||||||
| NC score | 0.004796 (rank : 95) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UIW0 | Gene names | VAX2 | |||
|
Domain Architecture |

|
|||||
| Description | Ventral anterior homeobox 2. | |||||
|
NRG2_HUMAN
|
||||||
| NC score | 0.004582 (rank : 96) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14511 | Gene names | NRG2, NTAK | |||
|
Domain Architecture |

|
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural- and thymus-derived activator for ERBB kinases) (NTAK) (Divergent of neuregulin-1) (DON-1)]. | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.003707 (rank : 97) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
VTNC_MOUSE
|
||||||
| NC score | 0.003662 (rank : 98) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.003184 (rank : 99) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.003125 (rank : 100) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.001563 (rank : 101) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.001178 (rank : 102) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
AMHR2_HUMAN
|
||||||
| NC score | -0.001423 (rank : 103) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16671, Q13762 | Gene names | AMHR2, AMHR | |||
|
Domain Architecture |

|
|||||
| Description | Anti-Muellerian hormone type-2 receptor precursor (EC 2.7.11.30) (Anti-Muellerian hormone type II receptor) (AMH type II receptor) (MIS type II receptor) (MISRII) (MRII). | |||||
|
AATK_MOUSE
|
||||||
| NC score | -0.002794 (rank : 104) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80YE4, O35211, Q3U2U5, Q3UHR8, Q66JN3, Q80YE3, Q8CB63, Q8CHE2 | Gene names | Aatk, Kiaa0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | -0.003270 (rank : 105) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
REST_MOUSE
|
||||||
| NC score | -0.004378 (rank : 106) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||