Please be patient as the page loads
|
VTNC_MOUSE
|
||||||
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
VTNC_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.988445 (rank : 2) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
VTNC_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 1.37539e-32 (rank : 3) | NC score | 0.535801 (rank : 45) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 8.91499e-32 (rank : 4) | NC score | 0.472680 (rank : 46) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MMP24_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 5) | NC score | 0.746378 (rank : 5) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y5R2, Q9H440 | Gene names | MMP24, MT5MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP). | |||||
|
MMP24_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 6) | NC score | 0.746259 (rank : 6) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9R0S2, Q9Z0J9 | Gene names | Mmp24, Mmp21, Mt5mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP) (MMP-21). | |||||
|
MMP16_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 7) | NC score | 0.748361 (rank : 3) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P51512, Q14824, Q52H48 | Gene names | MMP16, MMPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP) (MMP- X2). | |||||
|
MMP16_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 8) | NC score | 0.747995 (rank : 4) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WTR0, Q9ERT6 | Gene names | Mmp16 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP). | |||||
|
MMP15_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 9) | NC score | 0.743279 (rank : 9) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P51511, Q14111 | Gene names | MMP15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP) (SMCP- 2). | |||||
|
MMP15_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 10) | NC score | 0.741363 (rank : 10) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O54732 | Gene names | Mmp15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP). | |||||
|
MMP14_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 11) | NC score | 0.745857 (rank : 7) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P53690, O08645, O35369 | Gene names | Mmp14, Mtmmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP-X1) (MT-MMP). | |||||
|
MMP14_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 12) | NC score | 0.745811 (rank : 8) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P50281, Q92678 | Gene names | MMP14 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP- X1). | |||||
|
MMP17_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 13) | NC score | 0.739673 (rank : 11) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9ULZ9, Q14850 | Gene names | MMP17, MT4MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP11_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 14) | NC score | 0.733988 (rank : 14) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02853 | Gene names | Mmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP17_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 15) | NC score | 0.737794 (rank : 13) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9R0S3 | Gene names | Mmp17, Mt4mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP19_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 16) | NC score | 0.738177 (rank : 12) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JHI0, Q8C360 | Gene names | Mmp19, Rasi | |||
|
Domain Architecture |
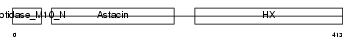
|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI). | |||||
|
MMP13_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 17) | NC score | 0.709050 (rank : 22) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P45452 | Gene names | MMP13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP19_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 18) | NC score | 0.732215 (rank : 16) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99542, O15278, O95606, Q99580 | Gene names | MMP19, MMP18, RASI | |||
|
Domain Architecture |
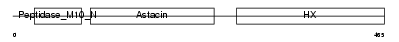
|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI) (MMP-18). | |||||
|
MMP8_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 19) | NC score | 0.711739 (rank : 19) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O70138, O88733 | Gene names | Mmp8 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (Collagenase 2). | |||||
|
MMP11_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 20) | NC score | 0.732637 (rank : 15) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP12_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 21) | NC score | 0.712583 (rank : 18) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P39900 | Gene names | MMP12, HME | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (HME) (Matrix metalloproteinase-12) (MMP-12) (Macrophage elastase) (ME). | |||||
|
MMP8_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 22) | NC score | 0.709711 (rank : 21) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P22894, Q45F99 | Gene names | MMP8, CLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (PMNL collagenase) (PMNL-CL). | |||||
|
MMP25_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 23) | NC score | 0.728896 (rank : 17) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NPA2, Q9H3Q0 | Gene names | MMP25, MT6MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-25 precursor (EC 3.4.24.-) (MMP-25) (Membrane-type matrix metalloproteinase 6) (MT-MMP 6) (Membrane-type-6 matrix metalloproteinase) (MT6-MMP) (Leukolysin). | |||||
|
MMP12_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 24) | NC score | 0.707971 (rank : 23) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P34960, Q8BJ92, Q8BJB3, Q8BJB6, Q8BJB8, Q8BJB9, Q8VED6 | Gene names | Mmp12, Mme, Mmel | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (MME) (Matrix metalloproteinase-12) (MMP-12). | |||||
|
HEMO_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 25) | NC score | 0.677717 (rank : 36) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P02790 | Gene names | HPX | |||
|
Domain Architecture |
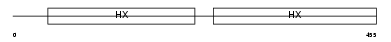
|
|||||
| Description | Hemopexin precursor (Beta-1B-glycoprotein). | |||||
|
MMP2_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 26) | NC score | 0.661829 (rank : 37) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P33434 | Gene names | Mmp2 | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A). | |||||
|
MMP2_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 27) | NC score | 0.660515 (rank : 38) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P08253 | Gene names | MMP2, CLG4A | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A) (TBE- 1). | |||||
|
MMP1_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 28) | NC score | 0.689871 (rank : 33) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P03956, P08156 | Gene names | MMP1, CLG | |||
|
Domain Architecture |

|
|||||
| Description | Interstitial collagenase precursor (EC 3.4.24.7) (Matrix metalloproteinase-1) (MMP-1) (Fibroblast collagenase) [Contains: 22 kDa interstitial collagenase; 27 kDa interstitial collagenase]. | |||||
|
MMP10_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 29) | NC score | 0.706111 (rank : 24) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O55123 | Gene names | Mmp10 | |||
|
Domain Architecture |
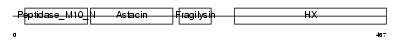
|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP1A_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 30) | NC score | 0.699528 (rank : 27) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9EPL5 | Gene names | Mmp1a, McolA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase A precursor (EC 3.4.24.7) (Matrix metalloproteinase-1a) (MMP-1a) (Mcol-A). | |||||
|
MMP3_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 31) | NC score | 0.697461 (rank : 29) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P28862 | Gene names | Mmp3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1) (EMS-2). | |||||
|
MMP3_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 32) | NC score | 0.699660 (rank : 26) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P08254, Q3B7S0 | Gene names | MMP3, STMY1 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1). | |||||
|
MMP1B_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 33) | NC score | 0.696686 (rank : 30) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9EPL6 | Gene names | Mmp1b, McolB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase B precursor (EC 3.4.24.7) (Matrix metalloproteinase-1b) (MMP-1b) (Mcol-B). | |||||
|
MMP10_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 34) | NC score | 0.701104 (rank : 25) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P09238 | Gene names | MMP10, STMY2 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP13_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 35) | NC score | 0.694723 (rank : 31) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P33435 | Gene names | Mmp13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP21_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 36) | NC score | 0.699118 (rank : 28) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K3F2 | Gene names | Mmp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
HEMO_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 37) | NC score | 0.599346 (rank : 43) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91X72, P97824, Q8WUP0 | Gene names | Hpx, Hpxn | |||
|
Domain Architecture |
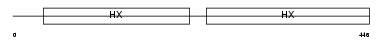
|
|||||
| Description | Hemopexin precursor. | |||||
|
MMP28_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 38) | NC score | 0.711425 (rank : 20) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9H239, Q96TE2 | Gene names | MMP28, MMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-28 precursor (EC 3.4.24.-) (MMP-28) (Epilysin). | |||||
|
MMP9_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 39) | NC score | 0.613358 (rank : 42) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
MMP21_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 40) | NC score | 0.690146 (rank : 32) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N119, Q8NG02 | Gene names | MMP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
MMP20_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 41) | NC score | 0.683440 (rank : 35) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P57748 | Gene names | Mmp20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP20_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 42) | NC score | 0.684767 (rank : 34) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O60882, Q6DKT9 | Gene names | MMP20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
ENPP2_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 43) | NC score | 0.166359 (rank : 48) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 44) | NC score | 0.165536 (rank : 49) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 45) | NC score | 0.154892 (rank : 50) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
MMP9_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 46) | NC score | 0.588614 (rank : 44) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
ENPP1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 47) | NC score | 0.154871 (rank : 51) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
PP11_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 48) | NC score | 0.253478 (rank : 47) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P21128, Q2NKJ4 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Placental protein 11 precursor (EC 3.4.21.-) (PP11). | |||||
|
SYNJ2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 49) | NC score | 0.030392 (rank : 73) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D2G5, O35404, O88399, O88400, O88401, O88402, O88403, O88404 | Gene names | Synj2, Kiaa0348 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
BCL7B_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 50) | NC score | 0.057028 (rank : 63) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921K9, O89022, Q3TV31, Q3U2W0 | Gene names | Bcl7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member B. | |||||
|
BCL7B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 51) | NC score | 0.052385 (rank : 67) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQE9, O43769, Q13845, Q6ZW75 | Gene names | BCL7B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member B. | |||||
|
ENPP3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.134652 (rank : 52) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.038429 (rank : 72) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LAMA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.004688 (rank : 86) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.024096 (rank : 74) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
G3BP2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.019218 (rank : 76) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97379, Q9R1B8 | Gene names | G3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
FAK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | -0.001457 (rank : 92) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14289, Q13475, Q14290, Q16709 | Gene names | PTK2B, FAK2, PYK2, RAFTK | |||
|
Domain Architecture |

|
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
NEK11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | -0.002715 (rank : 93) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NG66, Q5JPC0, Q8NG65, Q8TBY1, Q9H5F4 | Gene names | NEK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase Nek11 (EC 2.7.11.1) (NimA-related protein kinase 11) (Never in mitosis A-related kinase 11). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.005067 (rank : 85) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
TR10C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.012351 (rank : 81) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |
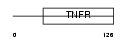
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
AQR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.019972 (rank : 75) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60306, Q2YDX9, Q6IRU8, Q6PIC8 | Gene names | AQR, KIAA0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius (Intron-binding protein of 160 kDa) (IBP160). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.005099 (rank : 84) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
FOXC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.003380 (rank : 88) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12948, Q86UP7, Q9BYM1, Q9NUE5, Q9UDD0, Q9UP06 | Gene names | FOXC1, FKHL7, FREAC3 | |||
|
Domain Architecture |
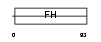
|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3). | |||||
|
G3BP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.015808 (rank : 78) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UN86, O60606, O75149, Q9UPA1 | Gene names | G3BP2, KIAA0660 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | -0.001082 (rank : 91) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
HA11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | -0.000278 (rank : 90) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01899 | Gene names | H2-D1 | |||
|
Domain Architecture |
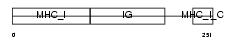
|
|||||
| Description | H-2 class I histocompatibility antigen, D-B alpha chain precursor (H- 2D(B)). | |||||
|
MADCA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.013609 (rank : 80) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.010407 (rank : 82) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
OCLN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.010342 (rank : 83) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16625, Q8N6K1 | Gene names | OCLN | |||
|
Domain Architecture |

|
|||||
| Description | Occludin. | |||||
|
SUSD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.041365 (rank : 71) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.017057 (rank : 77) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
ETV4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.003662 (rank : 87) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.014755 (rank : 79) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.002591 (rank : 89) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
ENPP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.061619 (rank : 59) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJA9, Q6UX49 | Gene names | ENPP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.061479 (rank : 60) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EQG7, Q921P7 | Gene names | Enpp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.059948 (rank : 61) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6UWR7, Q96M57 | Gene names | ENPP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.059657 (rank : 62) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BGN3, Q4VAH5 | Gene names | Enpp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.063317 (rank : 58) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.056526 (rank : 64) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MMP26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.619467 (rank : 41) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NRE1, Q9GZS2, Q9NR87 | Gene names | MMP26 | |||
|
Domain Architecture |
|
|||||
| Description | Matrix metalloproteinase-26 precursor (EC 3.4.24.-) (MMP-26) (Matrilysin-2) (Endometase). | |||||
|
MMP7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.621990 (rank : 39) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P09237, Q9BTK9 | Gene names | MMP7, MPSL1, PUMP1 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.619702 (rank : 40) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q10738 | Gene names | Mmp7 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
PRAX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.071281 (rank : 57) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PRAX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.056500 (rank : 65) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
SEL1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.052909 (rank : 66) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.051355 (rank : 69) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SPR1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.051753 (rank : 68) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.074004 (rank : 56) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SPR1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.050975 (rank : 70) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.079709 (rank : 55) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SPRR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.096610 (rank : 53) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
SPRR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.080258 (rank : 54) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
VTNC_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
VTNC_HUMAN
|
||||||
| NC score | 0.988445 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
MMP16_HUMAN
|
||||||
| NC score | 0.748361 (rank : 3) | θ value | 1.38178e-16 (rank : 7) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P51512, Q14824, Q52H48 | Gene names | MMP16, MMPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP) (MMP- X2). | |||||
|
MMP16_MOUSE
|
||||||
| NC score | 0.747995 (rank : 4) | θ value | 4.02038e-16 (rank : 8) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WTR0, Q9ERT6 | Gene names | Mmp16 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP). | |||||
|
MMP24_HUMAN
|
||||||
| NC score | 0.746378 (rank : 5) | θ value | 2.7842e-17 (rank : 5) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y5R2, Q9H440 | Gene names | MMP24, MT5MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP). | |||||
|
MMP24_MOUSE
|
||||||
| NC score | 0.746259 (rank : 6) | θ value | 1.058e-16 (rank : 6) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9R0S2, Q9Z0J9 | Gene names | Mmp24, Mmp21, Mt5mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP) (MMP-21). | |||||
|
MMP14_MOUSE
|
||||||
| NC score | 0.745857 (rank : 7) | θ value | 2.20605e-14 (rank : 11) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P53690, O08645, O35369 | Gene names | Mmp14, Mtmmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP-X1) (MT-MMP). | |||||
|
MMP14_HUMAN
|
||||||
| NC score | 0.745811 (rank : 8) | θ value | 2.88119e-14 (rank : 12) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P50281, Q92678 | Gene names | MMP14 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP- X1). | |||||
|
MMP15_HUMAN
|
||||||
| NC score | 0.743279 (rank : 9) | θ value | 5.25075e-16 (rank : 9) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P51511, Q14111 | Gene names | MMP15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP) (SMCP- 2). | |||||
|
MMP15_MOUSE
|
||||||
| NC score | 0.741363 (rank : 10) | θ value | 4.44505e-15 (rank : 10) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O54732 | Gene names | Mmp15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP). | |||||
|
MMP17_HUMAN
|
||||||
| NC score | 0.739673 (rank : 11) | θ value | 1.42992e-13 (rank : 13) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9ULZ9, Q14850 | Gene names | MMP17, MT4MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP19_MOUSE
|
||||||
| NC score | 0.738177 (rank : 12) | θ value | 1.02475e-11 (rank : 16) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JHI0, Q8C360 | Gene names | Mmp19, Rasi | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI). | |||||
|
MMP17_MOUSE
|
||||||
| NC score | 0.737794 (rank : 13) | θ value | 4.16044e-13 (rank : 15) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9R0S3 | Gene names | Mmp17, Mt4mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP11_MOUSE
|
||||||
| NC score | 0.733988 (rank : 14) | θ value | 3.18553e-13 (rank : 14) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02853 | Gene names | Mmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP11_HUMAN
|
||||||
| NC score | 0.732637 (rank : 15) | θ value | 1.133e-10 (rank : 20) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP19_HUMAN
|
||||||
| NC score | 0.732215 (rank : 16) | θ value | 2.98157e-11 (rank : 18) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99542, O15278, O95606, Q99580 | Gene names | MMP19, MMP18, RASI | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI) (MMP-18). | |||||
|
MMP25_HUMAN
|
||||||
| NC score | 0.728896 (rank : 17) | θ value | 3.29651e-10 (rank : 23) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NPA2, Q9H3Q0 | Gene names | MMP25, MT6MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-25 precursor (EC 3.4.24.-) (MMP-25) (Membrane-type matrix metalloproteinase 6) (MT-MMP 6) (Membrane-type-6 matrix metalloproteinase) (MT6-MMP) (Leukolysin). | |||||
|
MMP12_HUMAN
|
||||||
| NC score | 0.712583 (rank : 18) | θ value | 1.47974e-10 (rank : 21) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P39900 | Gene names | MMP12, HME | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (HME) (Matrix metalloproteinase-12) (MMP-12) (Macrophage elastase) (ME). | |||||
|
MMP8_MOUSE
|
||||||
| NC score | 0.711739 (rank : 19) | θ value | 6.64225e-11 (rank : 19) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O70138, O88733 | Gene names | Mmp8 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (Collagenase 2). | |||||
|
MMP28_HUMAN
|
||||||
| NC score | 0.711425 (rank : 20) | θ value | 9.92553e-07 (rank : 38) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9H239, Q96TE2 | Gene names | MMP28, MMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-28 precursor (EC 3.4.24.-) (MMP-28) (Epilysin). | |||||
|
MMP8_HUMAN
|
||||||
| NC score | 0.709711 (rank : 21) | θ value | 1.47974e-10 (rank : 22) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P22894, Q45F99 | Gene names | MMP8, CLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (PMNL collagenase) (PMNL-CL). | |||||
|
MMP13_HUMAN
|
||||||
| NC score | 0.709050 (rank : 22) | θ value | 2.98157e-11 (rank : 17) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P45452 | Gene names | MMP13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP12_MOUSE
|
||||||
| NC score | 0.707971 (rank : 23) | θ value | 1.25267e-09 (rank : 24) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P34960, Q8BJ92, Q8BJB3, Q8BJB6, Q8BJB8, Q8BJB9, Q8VED6 | Gene names | Mmp12, Mme, Mmel | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (MME) (Matrix metalloproteinase-12) (MMP-12). | |||||
|
MMP10_MOUSE
|
||||||
| NC score | 0.706111 (rank : 24) | θ value | 8.11959e-09 (rank : 29) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O55123 | Gene names | Mmp10 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP10_HUMAN
|
||||||
| NC score | 0.701104 (rank : 25) | θ value | 6.87365e-08 (rank : 34) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P09238 | Gene names | MMP10, STMY2 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP3_HUMAN
|
||||||
| NC score | 0.699660 (rank : 26) | θ value | 4.0297e-08 (rank : 32) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P08254, Q3B7S0 | Gene names | MMP3, STMY1 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1). | |||||
|
MMP1A_MOUSE
|
||||||
| NC score | 0.699528 (rank : 27) | θ value | 8.11959e-09 (rank : 30) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9EPL5 | Gene names | Mmp1a, McolA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase A precursor (EC 3.4.24.7) (Matrix metalloproteinase-1a) (MMP-1a) (Mcol-A). | |||||
|
MMP21_MOUSE
|
||||||
| NC score | 0.699118 (rank : 28) | θ value | 5.81887e-07 (rank : 36) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K3F2 | Gene names | Mmp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
MMP3_MOUSE
|
||||||
| NC score | 0.697461 (rank : 29) | θ value | 3.08544e-08 (rank : 31) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P28862 | Gene names | Mmp3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1) (EMS-2). | |||||
|
MMP1B_MOUSE
|
||||||
| NC score | 0.696686 (rank : 30) | θ value | 5.26297e-08 (rank : 33) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9EPL6 | Gene names | Mmp1b, McolB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase B precursor (EC 3.4.24.7) (Matrix metalloproteinase-1b) (MMP-1b) (Mcol-B). | |||||
|
MMP13_MOUSE
|
||||||
| NC score | 0.694723 (rank : 31) | θ value | 1.99992e-07 (rank : 35) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P33435 | Gene names | Mmp13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP21_HUMAN
|
||||||
| NC score | 0.690146 (rank : 32) | θ value | 4.92598e-06 (rank : 40) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N119, Q8NG02 | Gene names | MMP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
MMP1_HUMAN
|
||||||
| NC score | 0.689871 (rank : 33) | θ value | 2.79066e-09 (rank : 28) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P03956, P08156 | Gene names | MMP1, CLG | |||
|
Domain Architecture |

|
|||||
| Description | Interstitial collagenase precursor (EC 3.4.24.7) (Matrix metalloproteinase-1) (MMP-1) (Fibroblast collagenase) [Contains: 22 kDa interstitial collagenase; 27 kDa interstitial collagenase]. | |||||
|
MMP20_HUMAN
|
||||||
| NC score | 0.684767 (rank : 34) | θ value | 4.1701e-05 (rank : 42) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O60882, Q6DKT9 | Gene names | MMP20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP20_MOUSE
|
||||||
| NC score | 0.683440 (rank : 35) | θ value | 3.19293e-05 (rank : 41) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P57748 | Gene names | Mmp20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
HEMO_HUMAN
|
||||||
| NC score | 0.677717 (rank : 36) | θ value | 1.63604e-09 (rank : 25) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P02790 | Gene names | HPX | |||
|
Domain Architecture |

|
|||||
| Description | Hemopexin precursor (Beta-1B-glycoprotein). | |||||
|
MMP2_MOUSE
|
||||||
| NC score | 0.661829 (rank : 37) | θ value | 1.63604e-09 (rank : 26) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P33434 | Gene names | Mmp2 | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A). | |||||
|
MMP2_HUMAN
|
||||||
| NC score | 0.660515 (rank : 38) | θ value | 2.13673e-09 (rank : 27) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P08253 | Gene names | MMP2, CLG4A | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A) (TBE- 1). | |||||
|
MMP7_HUMAN
|
||||||
| NC score | 0.621990 (rank : 39) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P09237, Q9BTK9 | Gene names | MMP7, MPSL1, PUMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP7_MOUSE
|
||||||
| NC score | 0.619702 (rank : 40) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q10738 | Gene names | Mmp7 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP26_HUMAN
|
||||||
| NC score | 0.619467 (rank : 41) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NRE1, Q9GZS2, Q9NR87 | Gene names | MMP26 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-26 precursor (EC 3.4.24.-) (MMP-26) (Matrilysin-2) (Endometase). | |||||
|
MMP9_HUMAN
|
||||||
| NC score | 0.613358 (rank : 42) | θ value | 1.29631e-06 (rank : 39) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
HEMO_MOUSE
|
||||||
| NC score | 0.599346 (rank : 43) | θ value | 9.92553e-07 (rank : 37) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91X72, P97824, Q8WUP0 | Gene names | Hpx, Hpxn | |||
|
Domain Architecture |

|
|||||
| Description | Hemopexin precursor. | |||||
|
MMP9_MOUSE
|
||||||
| NC score | 0.588614 (rank : 44) | θ value | 0.00509761 (rank : 46) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.535801 (rank : 45) | θ value | 1.37539e-32 (rank : 3) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.472680 (rank : 46) | θ value | 8.91499e-32 (rank : 4) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PP11_HUMAN
|
||||||
| NC score | 0.253478 (rank : 47) | θ value | 0.0113563 (rank : 48) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P21128, Q2NKJ4 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Placental protein 11 precursor (EC 3.4.21.-) (PP11). | |||||
|
ENPP2_HUMAN
|
||||||
| NC score | 0.166359 (rank : 48) | θ value | 0.000602161 (rank : 43) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_MOUSE
|
||||||
| NC score | 0.165536 (rank : 49) | θ value | 0.00102713 (rank : 44) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_HUMAN
|
||||||
| NC score | 0.154892 (rank : 50) | θ value | 0.00390308 (rank : 45) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_MOUSE
|
||||||
| NC score | 0.154871 (rank : 51) | θ value | 0.00869519 (rank : 47) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP3_HUMAN
|
||||||
| NC score | 0.134652 (rank : 52) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
SPRR3_HUMAN
|
||||||
| NC score | 0.096610 (rank : 53) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
SPRR3_MOUSE
|
||||||
| NC score | 0.080258 (rank : 54) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.079709 (rank : 55) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.074004 (rank : 56) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.071281 (rank : 57) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
ENPP7_HUMAN
|
||||||
| NC score | 0.063317 (rank : 58) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
ENPP5_HUMAN
|
||||||
| NC score | 0.061619 (rank : 59) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJA9, Q6UX49 | Gene names | ENPP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP5_MOUSE
|
||||||
| NC score | 0.061479 (rank : 60) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EQG7, Q921P7 | Gene names | Enpp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP6_HUMAN
|
||||||
| NC score | 0.059948 (rank : 61) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6UWR7, Q96M57 | Gene names | ENPP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP6_MOUSE
|
||||||
| NC score | 0.059657 (rank : 62) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BGN3, Q4VAH5 | Gene names | Enpp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
BCL7B_MOUSE
|
||||||
| NC score | 0.057028 (rank : 63) | θ value | 0.0961366 (rank : 50) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921K9, O89022, Q3TV31, Q3U2W0 | Gene names | Bcl7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member B. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.056526 (rank : 64) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
PRAX_MOUSE
|
||||||
| NC score | 0.056500 (rank : 65) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
SEL1L_HUMAN
|
||||||
| NC score | 0.052909 (rank : 66) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
BCL7B_HUMAN
|
||||||
| NC score | 0.052385 (rank : 67) | θ value | 0.279714 (rank : 51) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQE9, O43769, Q13845, Q6ZW75 | Gene names | BCL7B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member B. | |||||
|
SPR1A_HUMAN
|
||||||
| NC score | 0.051753 (rank : 68) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
SEL1L_MOUSE
|
||||||
| NC score | 0.051355 (rank : 69) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SPR1B_HUMAN
|
||||||
| NC score | 0.050975 (rank : 70) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
SUSD2_HUMAN
|
||||||
| NC score | 0.041365 (rank : 71) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.038429 (rank : 72) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
SYNJ2_MOUSE
|
||||||
| NC score | 0.030392 (rank : 73) | θ value | 0.0431538 (rank : 49) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D2G5, O35404, O88399, O88400, O88401, O88402, O88403, O88404 | Gene names | Synj2, Kiaa0348 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.024096 (rank : 74) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
AQR_HUMAN
|
||||||
| NC score | 0.019972 (rank : 75) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60306, Q2YDX9, Q6IRU8, Q6PIC8 | Gene names | AQR, KIAA0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius (Intron-binding protein of 160 kDa) (IBP160). | |||||
|
G3BP2_MOUSE
|
||||||
| NC score | 0.019218 (rank : 76) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97379, Q9R1B8 | Gene names | G3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.017057 (rank : 77) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
G3BP2_HUMAN
|
||||||
| NC score | 0.015808 (rank : 78) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UN86, O60606, O75149, Q9UPA1 | Gene names | G3BP2, KIAA0660 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.014755 (rank : 79) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.013609 (rank : 80) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
TR10C_HUMAN
|
||||||
| NC score | 0.012351 (rank : 81) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.010407 (rank : 82) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
OCLN_HUMAN
|
||||||
| NC score | 0.010342 (rank : 83) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16625, Q8N6K1 | Gene names | OCLN | |||
|
Domain Architecture |

|
|||||
| Description | Occludin. | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.005099 (rank : 84) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.005067 (rank : 85) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
LAMA1_HUMAN
|
||||||
| NC score | 0.004688 (rank : 86) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
ETV4_MOUSE
|
||||||
| NC score | 0.003662 (rank : 87) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
FOXC1_HUMAN
|
||||||
| NC score | 0.003380 (rank : 88) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12948, Q86UP7, Q9BYM1, Q9NUE5, Q9UDD0, Q9UP06 | Gene names | FOXC1, FKHL7, FREAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.002591 (rank : 89) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
HA11_MOUSE
|
||||||
| NC score | -0.000278 (rank : 90) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01899 | Gene names | H2-D1 | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class I histocompatibility antigen, D-B alpha chain precursor (H- 2D(B)). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | -0.001082 (rank : 91) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
FAK2_HUMAN
|
||||||
| NC score | -0.001457 (rank : 92) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14289, Q13475, Q14290, Q16709 | Gene names | PTK2B, FAK2, PYK2, RAFTK | |||
|
Domain Architecture |

|
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
NEK11_HUMAN
|
||||||
| NC score | -0.002715 (rank : 93) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NG66, Q5JPC0, Q8NG65, Q8TBY1, Q9H5F4 | Gene names | NEK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase Nek11 (EC 2.7.11.1) (NimA-related protein kinase 11) (Never in mitosis A-related kinase 11). | |||||