Please be patient as the page loads
|
ELK1_MOUSE
|
||||||
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ELK1_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_HUMAN
|
||||||
| θ value | 2.85304e-163 (rank : 2) | NC score | 0.990370 (rank : 2) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK4_HUMAN
|
||||||
| θ value | 3.48141e-52 (rank : 3) | NC score | 0.948526 (rank : 6) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELK4_MOUSE
|
||||||
| θ value | 1.788e-48 (rank : 4) | NC score | 0.956594 (rank : 4) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELK3_HUMAN
|
||||||
| θ value | 2.50074e-42 (rank : 5) | NC score | 0.956722 (rank : 3) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELK3_MOUSE
|
||||||
| θ value | 1.2411e-41 (rank : 6) | NC score | 0.953667 (rank : 5) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ETV4_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 7) | NC score | 0.872708 (rank : 28) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV4_MOUSE
|
||||||
| θ value | 4.28545e-26 (rank : 8) | NC score | 0.873109 (rank : 27) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETV1_HUMAN
|
||||||
| θ value | 7.30988e-26 (rank : 9) | NC score | 0.875799 (rank : 26) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_MOUSE
|
||||||
| θ value | 7.30988e-26 (rank : 10) | NC score | 0.876682 (rank : 24) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV5_HUMAN
|
||||||
| θ value | 1.62847e-25 (rank : 11) | NC score | 0.870566 (rank : 29) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 1.62847e-25 (rank : 12) | NC score | 0.850899 (rank : 34) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ETS2_HUMAN
|
||||||
| θ value | 1.37858e-24 (rank : 13) | NC score | 0.896482 (rank : 9) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS2_MOUSE
|
||||||
| θ value | 1.37858e-24 (rank : 14) | NC score | 0.894982 (rank : 12) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETV3_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 15) | NC score | 0.875852 (rank : 25) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 16) | NC score | 0.850860 (rank : 35) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ERG_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 17) | NC score | 0.879598 (rank : 20) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ETS1_HUMAN
|
||||||
| θ value | 5.23862e-24 (rank : 18) | NC score | 0.895242 (rank : 10) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 19) | NC score | 0.895234 (rank : 11) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ERG_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 20) | NC score | 0.879676 (rank : 19) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
FLI1_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 21) | NC score | 0.884911 (rank : 16) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
FLI1_MOUSE
|
||||||
| θ value | 1.16704e-23 (rank : 22) | NC score | 0.886116 (rank : 15) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
ELF4_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 23) | NC score | 0.836139 (rank : 41) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ERF_HUMAN
|
||||||
| θ value | 6.40375e-22 (rank : 24) | NC score | 0.877486 (rank : 22) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| θ value | 6.40375e-22 (rank : 25) | NC score | 0.877157 (rank : 23) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ELF1_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 26) | NC score | 0.856675 (rank : 32) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
GABPA_HUMAN
|
||||||
| θ value | 7.0802e-21 (rank : 27) | NC score | 0.893468 (rank : 14) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
GABPA_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 28) | NC score | 0.894902 (rank : 13) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
ETV2_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 29) | NC score | 0.908849 (rank : 7) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 1.5773e-20 (rank : 30) | NC score | 0.848830 (rank : 37) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF4_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 31) | NC score | 0.856576 (rank : 33) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF2_MOUSE
|
||||||
| θ value | 5.99374e-20 (rank : 32) | NC score | 0.858986 (rank : 30) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 33) | NC score | 0.857518 (rank : 31) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ETV2_MOUSE
|
||||||
| θ value | 8.65492e-19 (rank : 34) | NC score | 0.904932 (rank : 8) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
SPDEF_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 35) | NC score | 0.880975 (rank : 18) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
SPDEF_MOUSE
|
||||||
| θ value | 9.56915e-18 (rank : 36) | NC score | 0.879391 (rank : 21) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ETV6_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 37) | NC score | 0.845715 (rank : 38) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 38) | NC score | 0.849211 (rank : 36) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV7_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 39) | NC score | 0.884425 (rank : 17) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ELF5_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 40) | NC score | 0.837785 (rank : 40) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF5_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 41) | NC score | 0.839051 (rank : 39) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
SPI1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 42) | NC score | 0.661005 (rank : 43) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPI1_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 43) | NC score | 0.662416 (rank : 42) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPIB_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 44) | NC score | 0.650220 (rank : 44) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIB_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 45) | NC score | 0.635013 (rank : 45) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIC_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 46) | NC score | 0.611928 (rank : 46) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 47) | NC score | 0.047697 (rank : 48) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SPIC_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 48) | NC score | 0.579281 (rank : 47) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
SYNP2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 49) | NC score | 0.039021 (rank : 50) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.032624 (rank : 52) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.030080 (rank : 53) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.020224 (rank : 67) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.019721 (rank : 68) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.026721 (rank : 56) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
TEX2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.022595 (rank : 63) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZPJ0 | Gene names | Tex2, Kiaa1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.002432 (rank : 100) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
AZI1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.002124 (rank : 101) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.024139 (rank : 59) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.023791 (rank : 60) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.025004 (rank : 57) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.019186 (rank : 69) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
GHR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.024446 (rank : 58) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10912, Q9HCX2 | Gene names | GHR | |||
|
Domain Architecture |
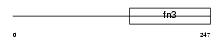
|
|||||
| Description | Growth hormone receptor precursor (GH receptor) (Somatotropin receptor) [Contains: Growth hormone-binding protein (GH-binding protein) (GHBP) (Serum-binding protein)]. | |||||
|
LIMD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.010617 (rank : 88) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
SIN3A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.017263 (rank : 75) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.026753 (rank : 55) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
K1958_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.015599 (rank : 80) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8K9, Q5T252, Q8TF43, Q96N02 | Gene names | KIAA1958 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958. | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.022152 (rank : 65) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
RP1L1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.017322 (rank : 73) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.006884 (rank : 95) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
GASP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.007807 (rank : 93) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
PHLB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.007327 (rank : 94) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.009534 (rank : 90) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | -0.002637 (rank : 107) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02086 | Gene names | SP2, KIAA0048 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp2. | |||||
|
GGA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.011724 (rank : 85) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.023203 (rank : 61) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MS3L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.017833 (rank : 70) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5Y2, Q9UG70, Q9Y5Z8 | Gene names | MSL3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
MS3L1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.017737 (rank : 72) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.005970 (rank : 98) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.016928 (rank : 77) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.017800 (rank : 71) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
RAI2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.022579 (rank : 64) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5P3, Q8N6X7 | Gene names | RAI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2. | |||||
|
SIGIR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.017115 (rank : 76) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLZ8, Q52L48 | Gene names | Sigirr, Tir8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Single Ig IL-1-related receptor (Single Ig IL-1R-related molecule) (Single immunoglobulin domain-containing IL1R-related protein) (Toll/interleukin-1 receptor 8) (TIR8). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.023186 (rank : 62) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.000154 (rank : 104) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
ASXL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.012987 (rank : 83) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.017304 (rank : 74) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
FOXM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.006331 (rank : 97) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08696 | Gene names | Foxm1, Fkh16 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead homolog 16) (Winged-helix transcription factor Trident). | |||||
|
HSPB9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.012947 (rank : 84) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DAM3 | Gene names | Hspb9 | |||
|
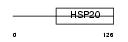
Domain Architecture |
|
|||||
| Description | Heat-shock protein beta-9 (HspB9). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.016447 (rank : 78) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
RAI2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.020607 (rank : 66) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QVY8 | Gene names | Rai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2 (3f8). | |||||
|
RIMB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.005262 (rank : 99) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15034, Q96ID2 | Gene names | RIMBP2, KIAA0318, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.011357 (rank : 86) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.029852 (rank : 54) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
TULP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.014313 (rank : 81) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JIL5, Q8CA75, Q922C2 | Gene names | Tulp4, Tusp | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
CO002_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.016444 (rank : 79) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
DUET_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | -0.005633 (rank : 110) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2A5, Q8TBQ5, Q9NSZ4 | Gene names | DUET, TRAD | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Duet (EC 2.7.11.1) (Serine/threonine kinase with Dbl- and pleckstrin homology domain). | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.010816 (rank : 87) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.006778 (rank : 96) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.039909 (rank : 49) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.002081 (rank : 102) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
TAF10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.034485 (rank : 51) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12962, O00703, Q13175 | Gene names | TAF10, TAF2H, TAFII30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 10 (Transcription initiation factor TFIID 30 kDa subunit) (TAF(II)30) (TAFII-30) (TAFII30) (STAF28). | |||||
|
ZN342_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | -0.003398 (rank : 108) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 705 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WUU4 | Gene names | ZNF342 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 342. | |||||
|
COAA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.009345 (rank : 91) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03692 | Gene names | COL10A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(X) chain precursor. | |||||
|
FREA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.001342 (rank : 103) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61574, Q8C5N7 | Gene names | Fkhl18, Fkh3, Freac10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein) (Transcription factor FKH-3). | |||||
|
HME1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | -0.001190 (rank : 106) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05925 | Gene names | EN1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein engrailed-1 (Hu-En-1). | |||||
|
RFIP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | -0.000488 (rank : 105) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BQP8, Q80T87 | Gene names | Rab11fip4, Kiaa1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
SIGIR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.010016 (rank : 89) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IA17, Q3KQY2, Q6UXI3, Q9H733 | Gene names | SIGIRR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Single Ig IL-1-related receptor (Single Ig IL-1R-related molecule) (Single immunoglobulin domain-containing IL1R-related protein) (Toll/interleukin-1 receptor 8) (TIR8). | |||||
|
SIN3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.013656 (rank : 82) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
ZBT16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | -0.005245 (rank : 109) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
ZSWM5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.008618 (rank : 92) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P217, Q5SXQ9 | Gene names | ZSWIM5, KIAA1511 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 5. | |||||
|
ELK1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_HUMAN
|
||||||
| NC score | 0.990370 (rank : 2) | θ value | 2.85304e-163 (rank : 2) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK3_HUMAN
|
||||||
| NC score | 0.956722 (rank : 3) | θ value | 2.50074e-42 (rank : 5) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELK4_MOUSE
|
||||||
| NC score | 0.956594 (rank : 4) | θ value | 1.788e-48 (rank : 4) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELK3_MOUSE
|
||||||
| NC score | 0.953667 (rank : 5) | θ value | 1.2411e-41 (rank : 6) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.948526 (rank : 6) | θ value | 3.48141e-52 (rank : 3) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ETV2_HUMAN
|
||||||
| NC score | 0.908849 (rank : 7) | θ value | 1.2077e-20 (rank : 29) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETV2_MOUSE
|
||||||
| NC score | 0.904932 (rank : 8) | θ value | 8.65492e-19 (rank : 34) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETS2_HUMAN
|
||||||
| NC score | 0.896482 (rank : 9) | θ value | 1.37858e-24 (rank : 13) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS1_HUMAN
|
||||||
| NC score | 0.895242 (rank : 10) | θ value | 5.23862e-24 (rank : 18) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| NC score | 0.895234 (rank : 11) | θ value | 5.23862e-24 (rank : 19) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS2_MOUSE
|
||||||
| NC score | 0.894982 (rank : 12) | θ value | 1.37858e-24 (rank : 14) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
GABPA_MOUSE
|
||||||
| NC score | 0.894902 (rank : 13) | θ value | 7.0802e-21 (rank : 28) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
GABPA_HUMAN
|
||||||
| NC score | 0.893468 (rank : 14) | θ value | 7.0802e-21 (rank : 27) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
FLI1_MOUSE
|
||||||
| NC score | 0.886116 (rank : 15) | θ value | 1.16704e-23 (rank : 22) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
FLI1_HUMAN
|
||||||
| NC score | 0.884911 (rank : 16) | θ value | 1.16704e-23 (rank : 21) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
ETV7_HUMAN
|
||||||
| NC score | 0.884425 (rank : 17) | θ value | 1.80466e-16 (rank : 39) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
SPDEF_HUMAN
|
||||||
| NC score | 0.880975 (rank : 18) | θ value | 7.32683e-18 (rank : 35) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ERG_HUMAN
|
||||||
| NC score | 0.879676 (rank : 19) | θ value | 8.93572e-24 (rank : 20) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
ERG_MOUSE
|
||||||
| NC score | 0.879598 (rank : 20) | θ value | 5.23862e-24 (rank : 17) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
SPDEF_MOUSE
|
||||||
| NC score | 0.879391 (rank : 21) | θ value | 9.56915e-18 (rank : 36) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ERF_HUMAN
|
||||||
| NC score | 0.877486 (rank : 22) | θ value | 6.40375e-22 (rank : 24) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| NC score | 0.877157 (rank : 23) | θ value | 6.40375e-22 (rank : 25) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ETV1_MOUSE
|
||||||
| NC score | 0.876682 (rank : 24) | θ value | 7.30988e-26 (rank : 10) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV3_MOUSE
|
||||||
| NC score | 0.875852 (rank : 25) | θ value | 3.07116e-24 (rank : 15) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV1_HUMAN
|
||||||
| NC score | 0.875799 (rank : 26) | θ value | 7.30988e-26 (rank : 9) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV4_MOUSE
|
||||||
| NC score | 0.873109 (rank : 27) | θ value | 4.28545e-26 (rank : 8) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETV4_HUMAN
|
||||||
| NC score | 0.872708 (rank : 28) | θ value | 4.28545e-26 (rank : 7) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.870566 (rank : 29) | θ value | 1.62847e-25 (rank : 11) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ELF2_MOUSE
|
||||||
| NC score | 0.858986 (rank : 30) | θ value | 5.99374e-20 (rank : 32) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_HUMAN
|
||||||
| NC score | 0.857518 (rank : 31) | θ value | 1.33526e-19 (rank : 33) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.856675 (rank : 32) | θ value | 4.15078e-21 (rank : 26) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF4_HUMAN
|
||||||
| NC score | 0.856576 (rank : 33) | θ value | 3.51386e-20 (rank : 31) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.850899 (rank : 34) | θ value | 1.62847e-25 (rank : 12) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.850860 (rank : 35) | θ value | 4.01107e-24 (rank : 16) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV6_MOUSE
|
||||||
| NC score | 0.849211 (rank : 36) | θ value | 1.63225e-17 (rank : 38) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.848830 (rank : 37) | θ value | 1.5773e-20 (rank : 30) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ETV6_HUMAN
|
||||||
| NC score | 0.845715 (rank : 38) | θ value | 1.63225e-17 (rank : 37) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ELF5_MOUSE
|
||||||
| NC score | 0.839051 (rank : 39) | θ value | 4.16044e-13 (rank : 41) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
ELF5_HUMAN
|
||||||
| NC score | 0.837785 (rank : 40) | θ value | 4.16044e-13 (rank : 40) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF4_MOUSE
|
||||||
| NC score | 0.836139 (rank : 41) | θ value | 3.75424e-22 (rank : 23) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
SPI1_MOUSE
|
||||||
| NC score | 0.662416 (rank : 42) | θ value | 4.45536e-07 (rank : 43) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPI1_HUMAN
|
||||||
| NC score | 0.661005 (rank : 43) | θ value | 4.45536e-07 (rank : 42) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPIB_HUMAN
|
||||||
| NC score | 0.650220 (rank : 44) | θ value | 1.09739e-05 (rank : 44) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIB_MOUSE
|
||||||
| NC score | 0.635013 (rank : 45) | θ value | 1.43324e-05 (rank : 45) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIC_HUMAN
|
||||||
| NC score | 0.611928 (rank : 46) | θ value | 7.1131e-05 (rank : 46) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPIC_MOUSE
|
||||||
| NC score | 0.579281 (rank : 47) | θ value | 0.000602161 (rank : 48) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.047697 (rank : 48) | θ value | 0.000270298 (rank : 47) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.039909 (rank : 49) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SYNP2_HUMAN
|
||||||
| NC score | 0.039021 (rank : 50) | θ value | 0.00134147 (rank : 49) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
TAF10_HUMAN
|
||||||
| NC score | 0.034485 (rank : 51) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12962, O00703, Q13175 | Gene names | TAF10, TAF2H, TAFII30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 10 (Transcription initiation factor TFIID 30 kDa subunit) (TAF(II)30) (TAFII-30) (TAFII30) (STAF28). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.032624 (rank : 52) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.030080 (rank : 53) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.029852 (rank : 54) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.026753 (rank : 55) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.026721 (rank : 56) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.025004 (rank : 57) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
GHR_HUMAN
|
||||||
| NC score | 0.024446 (rank : 58) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10912, Q9HCX2 | Gene names | GHR | |||
|
Domain Architecture |

|
|||||
| Description | Growth hormone receptor precursor (GH receptor) (Somatotropin receptor) [Contains: Growth hormone-binding protein (GH-binding protein) (GHBP) (Serum-binding protein)]. | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.024139 (rank : 59) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.023791 (rank : 60) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.023203 (rank : 61) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.023186 (rank : 62) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TEX2_MOUSE
|
||||||
| NC score | 0.022595 (rank : 63) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZPJ0 | Gene names | Tex2, Kiaa1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
RAI2_HUMAN
|
||||||
| NC score | 0.022579 (rank : 64) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5P3, Q8N6X7 | Gene names | RAI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2. | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.022152 (rank : 65) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
RAI2_MOUSE
|
||||||
| NC score | 0.020607 (rank : 66) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QVY8 | Gene names | Rai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2 (3f8). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.020224 (rank : 67) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.019721 (rank : 68) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.019186 (rank : 69) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MS3L1_HUMAN
|
||||||
| NC score | 0.017833 (rank : 70) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5Y2, Q9UG70, Q9Y5Z8 | Gene names | MSL3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.017800 (rank : 71) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MS3L1_MOUSE
|
||||||
| NC score | 0.017737 (rank : 72) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
RP1L1_MOUSE
|
||||||
| NC score | 0.017322 (rank : 73) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.017304 (rank : 74) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
SIN3A_HUMAN
|
||||||
| NC score | 0.017263 (rank : 75) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
SIGIR_MOUSE
|
||||||
| NC score | 0.017115 (rank : 76) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLZ8, Q52L48 | Gene names | Sigirr, Tir8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Single Ig IL-1-related receptor (Single Ig IL-1R-related molecule) (Single immunoglobulin domain-containing IL1R-related protein) (Toll/interleukin-1 receptor 8) (TIR8). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.016928 (rank : 77) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.016447 (rank : 78) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.016444 (rank : 79) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
K1958_HUMAN
|
||||||
| NC score | 0.015599 (rank : 80) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8K9, Q5T252, Q8TF43, Q96N02 | Gene names | KIAA1958 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958. | |||||
|
TULP4_MOUSE
|
||||||
| NC score | 0.014313 (rank : 81) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JIL5, Q8CA75, Q922C2 | Gene names | Tulp4, Tusp | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
SIN3A_MOUSE
|
||||||
| NC score | 0.013656 (rank : 82) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
ASXL1_HUMAN
|
||||||
| NC score | 0.012987 (rank : 83) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
HSPB9_MOUSE
|
||||||
| NC score | 0.012947 (rank : 84) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DAM3 | Gene names | Hspb9 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-9 (HspB9). | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.011724 (rank : 85) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.011357 (rank : 86) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.010816 (rank : 87) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
LIMD1_MOUSE
|
||||||
| NC score | 0.010617 (rank : 88) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
SIGIR_HUMAN
|
||||||
| NC score | 0.010016 (rank : 89) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IA17, Q3KQY2, Q6UXI3, Q9H733 | Gene names | SIGIRR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Single Ig IL-1-related receptor (Single Ig IL-1R-related molecule) (Single immunoglobulin domain-containing IL1R-related protein) (Toll/interleukin-1 receptor 8) (TIR8). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.009534 (rank : 90) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
COAA1_HUMAN
|
||||||
| NC score | 0.009345 (rank : 91) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03692 | Gene names | COL10A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(X) chain precursor. | |||||
|
ZSWM5_HUMAN
|
||||||
| NC score | 0.008618 (rank : 92) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P217, Q5SXQ9 | Gene names | ZSWIM5, KIAA1511 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 5. | |||||
|
GASP2_MOUSE
|
||||||
| NC score | 0.007807 (rank : 93) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
PHLB1_HUMAN
|
||||||
| NC score | 0.007327 (rank : 94) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.006884 (rank : 95) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.006778 (rank : 96) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
FOXM1_MOUSE
|
||||||
| NC score | 0.006331 (rank : 97) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08696 | Gene names | Foxm1, Fkh16 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead homolog 16) (Winged-helix transcription factor Trident). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.005970 (rank : 98) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
RIMB2_HUMAN
|
||||||
| NC score | 0.005262 (rank : 99) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15034, Q96ID2 | Gene names | RIMBP2, KIAA0318, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.002432 (rank : 100) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
AZI1_HUMAN
|
||||||
| NC score | 0.002124 (rank : 101) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.002081 (rank : 102) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
FREA_MOUSE
|
||||||
| NC score | 0.001342 (rank : 103) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61574, Q8C5N7 | Gene names | Fkhl18, Fkh3, Freac10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein) (Transcription factor FKH-3). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.000154 (rank : 104) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
RFIP4_MOUSE
|
||||||
| NC score | -0.000488 (rank : 105) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BQP8, Q80T87 | Gene names | Rab11fip4, Kiaa1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
HME1_HUMAN
|
||||||
| NC score | -0.001190 (rank : 106) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05925 | Gene names | EN1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein engrailed-1 (Hu-En-1). | |||||
|
SP2_HUMAN
|
||||||
| NC score | -0.002637 (rank : 107) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02086 | Gene names | SP2, KIAA0048 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp2. | |||||
|
ZN342_HUMAN
|
||||||
| NC score | -0.003398 (rank : 108) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 705 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WUU4 | Gene names | ZNF342 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 342. | |||||
|
ZBT16_HUMAN
|
||||||
| NC score | -0.005245 (rank : 109) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
DUET_HUMAN
|
||||||
| NC score | -0.005633 (rank : 110) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 110 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2A5, Q8TBQ5, Q9NSZ4 | Gene names | DUET, TRAD | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Duet (EC 2.7.11.1) (Serine/threonine kinase with Dbl- and pleckstrin homology domain). | |||||