Please be patient as the page loads
|
RP1L1_MOUSE
|
||||||
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.622350 (rank : 4) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
RP1L1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 170 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
RP1_MOUSE
|
||||||
| θ value | 3.37202e-47 (rank : 3) | NC score | 0.732604 (rank : 3) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |
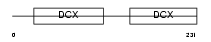
|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
RP1_HUMAN
|
||||||
| θ value | 2.18568e-46 (rank : 4) | NC score | 0.732644 (rank : 2) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P56715 | Gene names | RP1, ORP1 | |||
|
Domain Architecture |
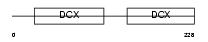
|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein) (Retinitis pigmentosa 1 protein). | |||||
|
DCAK1_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 5) | NC score | 0.144163 (rank : 9) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JLM8, Q6P207 | Gene names | Dcamkl1, Dclk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase DCAMKL1 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 1). | |||||
|
DCX_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 6) | NC score | 0.595913 (rank : 5) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
DCAK1_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 7) | NC score | 0.144046 (rank : 10) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 877 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15075 | Gene names | DCAMKL1, KIAA0369 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL1 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 1). | |||||
|
DCX_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 8) | NC score | 0.588293 (rank : 6) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |
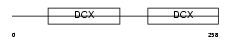
|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
DCAK2_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 9) | NC score | 0.140653 (rank : 11) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N568, Q8N399 | Gene names | DCAMKL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL2 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 2). | |||||
|
DCAK2_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 10) | NC score | 0.139217 (rank : 12) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PGN3, Q8BUU0, Q8BX25 | Gene names | Dcamkl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL2 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 2). | |||||
|
DCDC2_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 11) | NC score | 0.545975 (rank : 7) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |
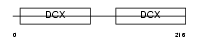
|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
DCDC2_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 12) | NC score | 0.513636 (rank : 8) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
CLIC6_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 13) | NC score | 0.064921 (rank : 25) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 14) | NC score | 0.075313 (rank : 18) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
HAIR_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 15) | NC score | 0.057264 (rank : 35) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
HS74L_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.025196 (rank : 111) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.052901 (rank : 44) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CS021_MOUSE
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.058340 (rank : 33) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D279, Q3UI04 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf21 homolog. | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.031748 (rank : 80) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
EDEM3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.050785 (rank : 53) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
|
KCTD9_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.047979 (rank : 58) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7L273, Q6NUM8, Q9NXV4 | Gene names | KCTD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
BASP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.085487 (rank : 14) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.072745 (rank : 19) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PARD3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.025144 (rank : 113) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
SFR16_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.060768 (rank : 31) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N2M8, O96026, Q6UW71, Q96DX2 | Gene names | SFRS16, SWAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2). | |||||
|
CDN1C_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.049024 (rank : 56) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
EOMES_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.017728 (rank : 138) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95936, Q8TAZ2, Q9UPM7 | Gene names | EOMES, TBR2 | |||
|
Domain Architecture |
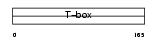
|
|||||
| Description | Eomesodermin homolog. | |||||
|
PER1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.029912 (rank : 91) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
AFF3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.032967 (rank : 76) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.027342 (rank : 100) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
KCTD9_MOUSE
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.043853 (rank : 60) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80UN1 | Gene names | Kctd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.051821 (rank : 48) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PCDH1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.006639 (rank : 184) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08174 | Gene names | PCDH1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-1 precursor (Protocadherin-42) (PC42) (Cadherin-like protein 1). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.050332 (rank : 55) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.033246 (rank : 74) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
ANXA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.014498 (rank : 148) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P12429, Q6LET2 | Gene names | ANXA3, ANX3 | |||
|
Domain Architecture |
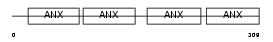
|
|||||
| Description | Annexin A3 (Annexin III) (Lipocortin III) (Placental anticoagulant protein III) (PAP-III) (35-alpha calcimedin) (Inositol 1,2-cyclic phosphate 2-phosphohydrolase). | |||||
|
CBX4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.027069 (rank : 102) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CD2L5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.024012 (rank : 117) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14004, Q6DKQ9, Q75MH4, Q75MH5, Q96JN4, Q9H4A0, Q9H4A1, Q9UDR4 | Gene names | CDC2L5, CDC2L, CHED, KIAA1791 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5) (Cholinesterase-related cell division controller). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.061497 (rank : 28) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
EURL_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.039082 (rank : 67) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D7G4 | Gene names | Eurl, D16Ertd472e | |||
|
Domain Architecture |

|
|||||
| Description | Protein EURL homolog. | |||||
|
ING1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.023337 (rank : 120) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXV3, Q9QUP8, Q9QXV4, Q9QZX3 | Gene names | Ing1 | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of growth protein 1. | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.030200 (rank : 87) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
LSD1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.030927 (rank : 84) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
OASL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.027982 (rank : 98) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15646, O75686, Q9Y6K6, Q9Y6K7 | Gene names | OASL, TRIP14 | |||
|
Domain Architecture |

|
|||||
| Description | 59 kDa 2'-5'-oligoadenylate synthetase-like protein (p59 OASL) (p59OASL) (Thyroid receptor-interacting protein 14) (TRIP14). | |||||
|
SEN34_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.042459 (rank : 62) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.031683 (rank : 81) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.021063 (rank : 129) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.048525 (rank : 57) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
COR2A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.012563 (rank : 152) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0P5, Q3U6H8, Q8BW42 | Gene names | Coro2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coronin-2A. | |||||
|
EXDL2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.031908 (rank : 78) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NVH0, Q8N3D3 | Gene names | EXDL2, C14orf114 | |||
|
Domain Architecture |
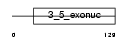
|
|||||
| Description | Exonuclease 3'-5' domain-like-containing protein 2. | |||||
|
GP73_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.061424 (rank : 29) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91XA2 | Gene names | Golph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
NETR_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.004982 (rank : 187) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08762 | Gene names | Prss12, Bssp3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Brain-specific serine protease 3) (BSSP-3). | |||||
|
PAK4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.030370 (rank : 86) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.039365 (rank : 65) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ZN263_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.001723 (rank : 191) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14978, O43387, Q96H95 | Gene names | ZNF263, FPM315 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 263 (Zinc finger protein FPM315). | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.011814 (rank : 157) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ZNF74_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.001244 (rank : 192) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16587, Q6IBV2, Q9UC04, Q9UF05, Q9UF06, Q9UF07 | Gene names | ZNF74 | |||
|
Domain Architecture |
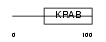
|
|||||
| Description | Zinc finger protein 74 (hZNF7). | |||||
|
CD2L5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.023285 (rank : 121) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
CENG3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.009776 (rank : 168) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
COBA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.028358 (rank : 95) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
EDD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.026618 (rank : 105) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
HIC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.003287 (rank : 189) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JLZ6, Q3U030, Q3ULP4, Q5K036, Q8BSZ9, Q8C3T5 | Gene names | Hic2, Kiaa1020 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 2 protein (Hic-2). | |||||
|
HXD11_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.007534 (rank : 180) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23813 | Gene names | Hoxd11, Hox-4.6, Hoxd-11 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D11 (Hox-4.6) (Hox-5.5). | |||||
|
JHD2A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.025106 (rank : 114) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.007248 (rank : 181) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.044048 (rank : 59) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
TBC10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.021161 (rank : 128) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58802 | Gene names | Tbc1d10a, Epi64, Tbc1d10 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 10A (EBP50-PDX interactor of 64 kDa) (EPI64 protein). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.025771 (rank : 108) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
ELK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.017322 (rank : 139) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
EMIL2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.016142 (rank : 145) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
GP124_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.011522 (rank : 158) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
JARD2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.022941 (rank : 122) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
KIFC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.009800 (rank : 167) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96AC6 | Gene names | KIFC2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC2. | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.036960 (rank : 68) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TSKS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.030858 (rank : 85) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UJT2, Q8WXJ0 | Gene names | STK22S1, TSKS, TSKS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine kinase substrate (Testis-specific kinase substrate) (STK22 substrate 1). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.043728 (rank : 61) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ZN516_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.011418 (rank : 159) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
CN118_MOUSE
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.028192 (rank : 97) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PE65, Q6ZPW9, Q8CD08, Q9DA49 | Gene names | Kiaa1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118 homolog. | |||||
|
COBA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.022593 (rank : 125) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.017209 (rank : 141) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
EDEM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.031189 (rank : 83) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92611 | Gene names | EDEM1, EDEM, KIAA0212 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.009717 (rank : 169) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
LPHN3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.008315 (rank : 174) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HAR2, O94867, Q9NWK5 | Gene names | LPHN3, KIAA0768, LEC3 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-3 precursor (Calcium-independent alpha-latrotoxin receptor 3) (Lectomedin-3). | |||||
|
NRG2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.016346 (rank : 144) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14511 | Gene names | NRG2, NTAK | |||
|
Domain Architecture |

|
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural- and thymus-derived activator for ERBB kinases) (NTAK) (Divergent of neuregulin-1) (DON-1)]. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.039809 (rank : 64) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.032616 (rank : 77) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.051754 (rank : 50) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
UBXD2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.031316 (rank : 82) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VCH8, Q8BW17 | Gene names | Ubxd2, Ubxdc1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.089682 (rank : 13) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.022042 (rank : 126) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.012549 (rank : 153) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
FOXN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.020391 (rank : 133) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.012349 (rank : 155) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.033681 (rank : 72) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.026738 (rank : 104) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.030128 (rank : 88) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.033098 (rank : 75) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.025156 (rank : 112) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.027095 (rank : 101) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.028192 (rank : 96) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
UBXD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.026924 (rank : 103) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.033698 (rank : 71) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.006927 (rank : 183) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
ARHGB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.012490 (rank : 154) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.050768 (rank : 54) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.029596 (rank : 92) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
CMGA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.033592 (rank : 73) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.010281 (rank : 163) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
EDEM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.028875 (rank : 94) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q925U4 | Gene names | Edem1, Edem | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.069929 (rank : 21) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
GET1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.022778 (rank : 123) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C7E7 | Gene names | D5Ertd593e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Genethonin-1. | |||||
|
JIP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.026453 (rank : 106) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.024819 (rank : 115) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.025762 (rank : 109) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.005226 (rank : 186) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
MKL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.013332 (rank : 150) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
PSD12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.016921 (rank : 143) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00232, Q53HA2, Q6P053 | Gene names | PSMD12 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 12 (26S proteasome regulatory subunit p55). | |||||
|
TCAL4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.020932 (rank : 132) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96EI5, Q8WY12, Q9H2H1, Q9H775 | Gene names | TCEAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 4 (TCEA-like protein 4) (Transcription elongation factor S-II protein-like 4). | |||||
|
TNR4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.014692 (rank : 147) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43489, Q13663, Q5T7M0 | Gene names | TNFRSF4, TXGP1L | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (ACT35 antigen) (TAX transcriptionally-activated glycoprotein 1 receptor) (CD134 antigen). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.024450 (rank : 116) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
UNC5B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.008307 (rank : 175) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IZJ1, Q86SN3, Q8N1Y2, Q9H9F3 | Gene names | UNC5B, P53RDL1, UNC5H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2) (p53-regulated receptor for death and life protein 1). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.039256 (rank : 66) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
ZN192_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.000460 (rank : 193) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15776, Q4VAR1, Q4VAR2, Q4VAR3, Q9H4T1 | Gene names | ZNF192 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 192 (LD5-1). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.007644 (rank : 179) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.011232 (rank : 160) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
APBA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.010763 (rank : 162) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.025360 (rank : 110) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
CDSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.027880 (rank : 99) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
CHFR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.009618 (rank : 170) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96EP1, Q96SL3, Q9NRT4, Q9NT32, Q9NVD5 | Gene names | CHFR | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase CHFR (EC 6.3.2.-) (Checkpoint with forkhead and RING finger domains protein). | |||||
|
CN118_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.023718 (rank : 119) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NWQ4, Q6PEJ7, Q9NWH0, Q9ULR8 | Gene names | C14orf118, KIAA1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118. | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.008987 (rank : 173) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.066861 (rank : 23) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
FRAT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.015565 (rank : 146) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K025 | Gene names | Frat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GSK-3-binding protein FRAT2 (Frat-2). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.034463 (rank : 70) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
IRS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.012212 (rank : 156) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35568 | Gene names | IRS1 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.010931 (rank : 161) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
RCC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.009881 (rank : 166) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P258, Q8IVL9, Q9BSN6, Q9NPV8 | Gene names | RCC2, KIAA1470, TD60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD- 60). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.040329 (rank : 63) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.021026 (rank : 130) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
RNH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.017814 (rank : 137) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70338, Q8VCR6 | Gene names | Rnaseh1, Rnh1 | |||
|
Domain Architecture |
|
|||||
| Description | Ribonuclease H1 (EC 3.1.26.4) (RNase H1). | |||||
|
SN1L2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.029440 (rank : 93) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CFH6 | Gene names | Snf1lk2, Sik2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Salt-inducible kinase 2). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.079551 (rank : 15) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
UHRF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.009000 (rank : 172) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
BRCA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.017295 (rank : 140) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.017096 (rank : 142) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CO027_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.014342 (rank : 149) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZB3, Q6NVG3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf27 homolog. | |||||
|
CO1A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.026333 (rank : 107) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CTDP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.035557 (rank : 69) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TSG2, Q7TSS7, Q9D4S8 | Gene names | Ctdp1, Fcp1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
CUTL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.018196 (rank : 136) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
CXA12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.004558 (rank : 188) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BQU6, Q6TLV2, Q8BQS2, Q9EPM1 | Gene names | Gja12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gap junction alpha-12 protein (Connexin-47) (Cx47). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.007786 (rank : 176) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
DMN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.030053 (rank : 90) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15061 | Gene names | DMN, KIAA0353 | |||
|
Domain Architecture |

|
|||||
| Description | Desmuslin. | |||||
|
ESCO1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.009935 (rank : 164) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q69Z69, Q8BQI2, Q8BR47, Q922F5 | Gene names | Esco1, Kiaa1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1). | |||||
|
EVL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.009556 (rank : 171) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
FOXN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.009911 (rank : 165) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.023788 (rank : 118) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.030106 (rank : 89) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
JAD1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.007717 (rank : 178) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.021283 (rank : 127) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MRP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.018636 (rank : 134) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.031845 (rank : 79) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.071471 (rank : 20) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
PER1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.020990 (rank : 131) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.018608 (rank : 135) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.002682 (rank : 190) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
PI51C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.022615 (rank : 124) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
RHG06_MOUSE
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.005741 (rank : 185) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
ROBO3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.007083 (rank : 182) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
ROBO3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.007731 (rank : 177) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2I4 | Gene names | Robo3, Rbig1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Retinoblastoma-inhibiting gene 1) (Rig-1). | |||||
|
ZN622_MOUSE
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.013028 (rank : 151) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VY9 | Gene names | Znf622, D15Ertd806e, Zfp622 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 622. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.064455 (rank : 26) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CABYR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.057633 (rank : 34) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75952, Q8WXW5, Q9HAY3, Q9HAY4, Q9HAY5, Q9HCY9 | Gene names | CABYR, CBP86, FSP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86) (Fibrousheathin-2) (FSP-2). | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.054137 (rank : 42) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.075326 (rank : 17) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CGRE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.058445 (rank : 32) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.066303 (rank : 24) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.076527 (rank : 16) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.057202 (rank : 36) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.055367 (rank : 38) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.054949 (rank : 41) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.052134 (rank : 47) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.060952 (rank : 30) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.055602 (rank : 37) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.051773 (rank : 49) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.052164 (rank : 46) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.052843 (rank : 45) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.068445 (rank : 22) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.054986 (rank : 40) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TARA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.053521 (rank : 43) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.055112 (rank : 39) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.051682 (rank : 51) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
TOIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.051165 (rank : 52) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.062513 (rank : 27) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
RP1L1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 170 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
RP1_HUMAN
|
||||||
| NC score | 0.732644 (rank : 2) | θ value | 2.18568e-46 (rank : 4) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P56715 | Gene names | RP1, ORP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein) (Retinitis pigmentosa 1 protein). | |||||
|
RP1_MOUSE
|
||||||
| NC score | 0.732604 (rank : 3) | θ value | 3.37202e-47 (rank : 3) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |

|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.622350 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
DCX_MOUSE
|
||||||
| NC score | 0.595913 (rank : 5) | θ value | 8.10077e-17 (rank : 6) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
DCX_HUMAN
|
||||||
| NC score | 0.588293 (rank : 6) | θ value | 2.60593e-15 (rank : 8) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
DCDC2_HUMAN
|
||||||
| NC score | 0.545975 (rank : 7) | θ value | 3.29651e-10 (rank : 11) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |

|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
DCDC2_MOUSE
|
||||||
| NC score | 0.513636 (rank : 8) | θ value | 8.11959e-09 (rank : 12) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
DCAK1_MOUSE
|
||||||
| NC score | 0.144163 (rank : 9) | θ value | 8.10077e-17 (rank : 5) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JLM8, Q6P207 | Gene names | Dcamkl1, Dclk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase DCAMKL1 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 1). | |||||
|
DCAK1_HUMAN
|
||||||
| NC score | 0.144046 (rank : 10) | θ value | 1.38178e-16 (rank : 7) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 877 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15075 | Gene names | DCAMKL1, KIAA0369 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL1 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 1). | |||||
|
DCAK2_HUMAN
|
||||||
| NC score | 0.140653 (rank : 11) | θ value | 6.41864e-14 (rank : 9) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N568, Q8N399 | Gene names | DCAMKL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL2 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 2). | |||||
|
DCAK2_MOUSE
|
||||||
| NC score | 0.139217 (rank : 12) | θ value | 1.42992e-13 (rank : 10) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PGN3, Q8BUU0, Q8BX25 | Gene names | Dcamkl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL2 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 2). | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.089682 (rank : 13) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.085487 (rank : 14) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.079551 (rank : 15) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.076527 (rank : 16) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.075326 (rank : 17) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.075313 (rank : 18) | θ value | 0.0193708 (rank : 14) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.072745 (rank : 19) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.071471 (rank : 20) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.069929 (rank : 21) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.068445 (rank : 22) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.066861 (rank : 23) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.066303 (rank : 24) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CLIC6_MOUSE
|
||||||
| NC score | 0.064921 (rank : 25) | θ value | 0.00134147 (rank : 13) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.064455 (rank : 26) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.062513 (rank : 27) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.061497 (rank : 28) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
GP73_MOUSE
|
||||||
| NC score | 0.061424 (rank : 29) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91XA2 | Gene names | Golph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.060952 (rank : 30) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SFR16_HUMAN
|
||||||
| NC score | 0.060768 (rank : 31) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N2M8, O96026, Q6UW71, Q96DX2 | Gene names | SFRS16, SWAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2). | |||||
|
CGRE1_MOUSE
|
||||||
| NC score | 0.058445 (rank : 32) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CS021_MOUSE
|
||||||
| NC score | 0.058340 (rank : 33) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D279, Q3UI04 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf21 homolog. | |||||
|
CABYR_HUMAN
|
||||||
| NC score | 0.057633 (rank : 34) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75952, Q8WXW5, Q9HAY3, Q9HAY4, Q9HAY5, Q9HCY9 | Gene names | CABYR, CBP86, FSP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86) (Fibrousheathin-2) (FSP-2). | |||||
|
HAIR_MOUSE
|
||||||
| NC score | 0.057264 (rank : 35) | θ value | 0.0193708 (rank : 15) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.057202 (rank : 36) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.055602 (rank : 37) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.055367 (rank : 38) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.055112 (rank : 39) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.054986 (rank : 40) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.054949 (rank : 41) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | 0.054137 (rank : 42) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.053521 (rank : 43) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.052901 (rank : 44) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PRPE_HUMAN
|
||||||
| NC score | 0.052843 (rank : 45) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.052164 (rank : 46) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.052134 (rank : 47) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.051821 (rank : 48) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.051773 (rank : 49) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.051754 (rank : 50) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.051682 (rank : 51) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
TOIP2_MOUSE
|
||||||
| NC score | 0.051165 (rank : 52) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
EDEM3_HUMAN
|
||||||
| NC score | 0.050785 (rank : 53) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.050768 (rank : 54) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.050332 (rank : 55) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
CDN1C_MOUSE
|
||||||
| NC score | 0.049024 (rank : 56) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.048525 (rank : 57) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
KCTD9_HUMAN
|
||||||
| NC score | 0.047979 (rank : 58) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7L273, Q6NUM8, Q9NXV4 | Gene names | KCTD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.044048 (rank : 59) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
KCTD9_MOUSE
|
||||||
| NC score | 0.043853 (rank : 60) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80UN1 | Gene names | Kctd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.043728 (rank : 61) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
SEN34_MOUSE
|
||||||
| NC score | 0.042459 (rank : 62) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.040329 (rank : 63) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.039809 (rank : 64) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.039365 (rank : 65) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.039256 (rank : 66) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
EURL_MOUSE
|
||||||
| NC score | 0.039082 (rank : 67) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D7G4 | Gene names | Eurl, D16Ertd472e | |||
|
Domain Architecture |

|
|||||
| Description | Protein EURL homolog. | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.036960 (rank : 68) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CTDP1_MOUSE
|
||||||
| NC score | 0.035557 (rank : 69) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TSG2, Q7TSS7, Q9D4S8 | Gene names | Ctdp1, Fcp1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.034463 (rank : 70) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.033698 (rank : 71) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.033681 (rank : 72) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
CMGA_MOUSE
|
||||||
| NC score | 0.033592 (rank : 73) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.033246 (rank : 74) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.033098 (rank : 75) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.032967 (rank : 76) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.032616 (rank : 77) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
EXDL2_HUMAN
|
||||||
| NC score | 0.031908 (rank : 78) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NVH0, Q8N3D3 | Gene names | EXDL2, C14orf114 | |||
|
Domain Architecture |
|
|||||
| Description | Exonuclease 3'-5' domain-like-containing protein 2. | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.031845 (rank : 79) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.031748 (rank : 80) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.031683 (rank : 81) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
UBXD2_MOUSE
|
||||||
| NC score | 0.031316 (rank : 82) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VCH8, Q8BW17 | Gene names | Ubxd2, Ubxdc1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
EDEM1_HUMAN
|
||||||
| NC score | 0.031189 (rank : 83) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92611 | Gene names | EDEM1, EDEM, KIAA0212 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
LSD1_MOUSE
|
||||||
| NC score | 0.030927 (rank : 84) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
TSKS_HUMAN
|
||||||
| NC score | 0.030858 (rank : 85) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UJT2, Q8WXJ0 | Gene names | STK22S1, TSKS, TSKS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine kinase substrate (Testis-specific kinase substrate) (STK22 substrate 1). | |||||
|
PAK4_MOUSE
|
||||||
| NC score | 0.030370 (rank : 86) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.030200 (rank : 87) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.030128 (rank : 88) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.030106 (rank : 89) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
DMN_HUMAN
|
||||||
| NC score | 0.030053 (rank : 90) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15061 | Gene names | DMN, KIAA0353 | |||
|
Domain Architecture |

|
|||||
| Description | Desmuslin. | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.029912 (rank : 91) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.029596 (rank : 92) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
SN1L2_MOUSE
|
||||||
| NC score | 0.029440 (rank : 93) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CFH6 | Gene names | Snf1lk2, Sik2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Salt-inducible kinase 2). | |||||
|
EDEM1_MOUSE
|
||||||
| NC score | 0.028875 (rank : 94) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q925U4 | Gene names | Edem1, Edem | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
COBA2_HUMAN
|
||||||
| NC score | 0.028358 (rank : 95) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.028192 (rank : 96) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
CN118_MOUSE
|
||||||
| NC score | 0.028192 (rank : 97) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PE65, Q6ZPW9, Q8CD08, Q9DA49 | Gene names | Kiaa1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118 homolog. | |||||
|
OASL_HUMAN
|
||||||
| NC score | 0.027982 (rank : 98) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15646, O75686, Q9Y6K6, Q9Y6K7 | Gene names | OASL, TRIP14 | |||
|
Domain Architecture |

|
|||||
| Description | 59 kDa 2'-5'-oligoadenylate synthetase-like protein (p59 OASL) (p59OASL) (Thyroid receptor-interacting protein 14) (TRIP14). | |||||
|
CDSN_MOUSE
|
||||||
| NC score | 0.027880 (rank : 99) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.027342 (rank : 100) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.027095 (rank : 101) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
CBX4_MOUSE
|
||||||
| NC score | 0.027069 (rank : 102) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
UBXD2_HUMAN
|
||||||
| NC score | 0.026924 (rank : 103) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.026738 (rank : 104) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.026618 (rank : 105) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
JIP4_MOUSE
|
||||||
| NC score | 0.026453 (rank : 106) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
CO1A1_HUMAN
|
||||||
| NC score | 0.026333 (rank : 107) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.025771 (rank : 108) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.025762 (rank : 109) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.025360 (rank : 110) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
HS74L_HUMAN
|
||||||
| NC score | 0.025196 (rank : 111) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.025156 (rank : 112) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PARD3_MOUSE
|
||||||
| NC score | 0.025144 (rank : 113) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
JHD2A_HUMAN
|
||||||
| NC score | 0.025106 (rank : 114) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.024819 (rank : 115) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.024450 (rank : 116) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
CD2L5_HUMAN
|
||||||
| NC score | 0.024012 (rank : 117) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14004, Q6DKQ9, Q75MH4, Q75MH5, Q96JN4, Q9H4A0, Q9H4A1, Q9UDR4 | Gene names | CDC2L5, CDC2L, CHED, KIAA1791 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5) (Cholinesterase-related cell division controller). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.023788 (rank : 118) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
CN118_HUMAN
|
||||||
| NC score | 0.023718 (rank : 119) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NWQ4, Q6PEJ7, Q9NWH0, Q9ULR8 | Gene names | C14orf118, KIAA1152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf118. | |||||
|
ING1_MOUSE
|
||||||
| NC score | 0.023337 (rank : 120) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXV3, Q9QUP8, Q9QXV4, Q9QZX3 | Gene names | Ing1 | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of growth protein 1. | |||||
|
CD2L5_MOUSE
|
||||||
| NC score | 0.023285 (rank : 121) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
JARD2_MOUSE
|
||||||
| NC score | 0.022941 (rank : 122) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
GET1_MOUSE
|
||||||
| NC score | 0.022778 (rank : 123) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C7E7 | Gene names | D5Ertd593e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Genethonin-1. | |||||
|
PI51C_HUMAN
|
||||||
| NC score | 0.022615 (rank : 124) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
COBA2_MOUSE
|
||||||
| NC score | 0.022593 (rank : 125) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.022042 (rank : 126) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.021283 (rank : 127) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
TBC10_MOUSE
|
||||||
| NC score | 0.021161 (rank : 128) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58802 | Gene names | Tbc1d10a, Epi64, Tbc1d10 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 10A (EBP50-PDX interactor of 64 kDa) (EPI64 protein). | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.021063 (rank : 129) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.021026 (rank : 130) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.020990 (rank : 131) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
TCAL4_HUMAN
|
||||||
| NC score | 0.020932 (rank : 132) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96EI5, Q8WY12, Q9H2H1, Q9H775 | Gene names | TCEAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 4 (TCEA-like protein 4) (Transcription elongation factor S-II protein-like 4). | |||||
|
FOXN1_HUMAN
|
||||||
| NC score | 0.020391 (rank : 133) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
MRP_MOUSE
|
||||||
| NC score | 0.018636 (rank : 134) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.018608 (rank : 135) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CUTL2_MOUSE
|
||||||
| NC score | 0.018196 (rank : 136) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
RNH1_MOUSE
|
||||||
| NC score | 0.017814 (rank : 137) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70338, Q8VCR6 | Gene names | Rnaseh1, Rnh1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease H1 (EC 3.1.26.4) (RNase H1). | |||||
|
EOMES_HUMAN
|
||||||
| NC score | 0.017728 (rank : 138) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95936, Q8TAZ2, Q9UPM7 | Gene names | EOMES, TBR2 | |||
|
Domain Architecture |

|
|||||
| Description | Eomesodermin homolog. | |||||
|
ELK1_MOUSE
|
||||||
| NC score | 0.017322 (rank : 139) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
BRCA2_HUMAN
|
||||||
| NC score | 0.017295 (rank : 140) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.017209 (rank : 141) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.017096 (rank : 142) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
PSD12_HUMAN
|
||||||
| NC score | 0.016921 (rank : 143) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00232, Q53HA2, Q6P053 | Gene names | PSMD12 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 12 (26S proteasome regulatory subunit p55). | |||||
|
NRG2_HUMAN
|
||||||
| NC score | 0.016346 (rank : 144) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14511 | Gene names | NRG2, NTAK | |||
|
Domain Architecture |

|
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural- and thymus-derived activator for ERBB kinases) (NTAK) (Divergent of neuregulin-1) (DON-1)]. | |||||
|
EMIL2_MOUSE
|
||||||
| NC score | 0.016142 (rank : 145) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
FRAT2_MOUSE
|
||||||
| NC score | 0.015565 (rank : 146) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K025 | Gene names | Frat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GSK-3-binding protein FRAT2 (Frat-2). | |||||
|
TNR4_HUMAN
|
||||||
| NC score | 0.014692 (rank : 147) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43489, Q13663, Q5T7M0 | Gene names | TNFRSF4, TXGP1L | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (ACT35 antigen) (TAX transcriptionally-activated glycoprotein 1 receptor) (CD134 antigen). | |||||
|
ANXA3_HUMAN
|
||||||
| NC score | 0.014498 (rank : 148) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P12429, Q6LET2 | Gene names | ANXA3, ANX3 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A3 (Annexin III) (Lipocortin III) (Placental anticoagulant protein III) (PAP-III) (35-alpha calcimedin) (Inositol 1,2-cyclic phosphate 2-phosphohydrolase). | |||||
|
CO027_MOUSE
|
||||||
| NC score | 0.014342 (rank : 149) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZB3, Q6NVG3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf27 homolog. | |||||
|
MKL1_HUMAN
|
||||||
| NC score | 0.013332 (rank : 150) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
ZN622_MOUSE
|
||||||
| NC score | 0.013028 (rank : 151) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VY9 | Gene names | Znf622, D15Ertd806e, Zfp622 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 622. | |||||
|
COR2A_MOUSE
|
||||||
| NC score | 0.012563 (rank : 152) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0P5, Q3U6H8, Q8BW42 | Gene names | Coro2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coronin-2A. | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.012549 (rank : 153) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
ARHGB_HUMAN
|
||||||
| NC score | 0.012490 (rank : 154) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.012349 (rank : 155) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
IRS1_HUMAN
|
||||||
| NC score | 0.012212 (rank : 156) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35568 | Gene names | IRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
ZN687_MOUSE
|
||||||
| NC score | 0.011814 (rank : 157) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
GP124_MOUSE
|
||||||
| NC score | 0.011522 (rank : 158) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
ZN516_HUMAN
|
||||||
| NC score | 0.011418 (rank : 159) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.011232 (rank : 160) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.010931 (rank : 161) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
APBA2_HUMAN
|
||||||
| NC score | 0.010763 (rank : 162) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.010281 (rank : 163) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ESCO1_MOUSE
|
||||||
| NC score | 0.009935 (rank : 164) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q69Z69, Q8BQI2, Q8BR47, Q922F5 | Gene names | Esco1, Kiaa1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1). | |||||
|
FOXN1_MOUSE
|
||||||
| NC score | 0.009911 (rank : 165) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
RCC2_HUMAN
|
||||||
| NC score | 0.009881 (rank : 166) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P258, Q8IVL9, Q9BSN6, Q9NPV8 | Gene names | RCC2, KIAA1470, TD60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD- 60). | |||||
|
KIFC2_HUMAN
|
||||||
| NC score | 0.009800 (rank : 167) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96AC6 | Gene names | KIFC2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC2. | |||||
|
CENG3_HUMAN
|
||||||
| NC score | 0.009776 (rank : 168) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.009717 (rank : 169) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
CHFR_HUMAN
|
||||||
| NC score | 0.009618 (rank : 170) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96EP1, Q96SL3, Q9NRT4, Q9NT32, Q9NVD5 | Gene names | CHFR | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase CHFR (EC 6.3.2.-) (Checkpoint with forkhead and RING finger domains protein). | |||||
|
EVL_HUMAN
|
||||||
| NC score | 0.009556 (rank : 171) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
UHRF1_MOUSE
|
||||||
| NC score | 0.009000 (rank : 172) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.008987 (rank : 173) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
LPHN3_HUMAN
|
||||||
| NC score | 0.008315 (rank : 174) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HAR2, O94867, Q9NWK5 | Gene names | LPHN3, KIAA0768, LEC3 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-3 precursor (Calcium-independent alpha-latrotoxin receptor 3) (Lectomedin-3). | |||||
|
UNC5B_HUMAN
|
||||||
| NC score | 0.008307 (rank : 175) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IZJ1, Q86SN3, Q8N1Y2, Q9H9F3 | Gene names | UNC5B, P53RDL1, UNC5H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2) (p53-regulated receptor for death and life protein 1). | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.007786 (rank : 176) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
ROBO3_MOUSE
|
||||||
| NC score | 0.007731 (rank : 177) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2I4 | Gene names | Robo3, Rbig1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Retinoblastoma-inhibiting gene 1) (Rig-1). | |||||
|
JAD1C_MOUSE
|
||||||
| NC score | 0.007717 (rank : 178) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.007644 (rank : 179) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
HXD11_MOUSE
|
||||||
| NC score | 0.007534 (rank : 180) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23813 | Gene names | Hoxd11, Hox-4.6, Hoxd-11 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D11 (Hox-4.6) (Hox-5.5). | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.007248 (rank : 181) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
ROBO3_HUMAN
|
||||||
| NC score | 0.007083 (rank : 182) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.006927 (rank : 183) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
PCDH1_HUMAN
|
||||||
| NC score | 0.006639 (rank : 184) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08174 | Gene names | PCDH1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-1 precursor (Protocadherin-42) (PC42) (Cadherin-like protein 1). | |||||
|
RHG06_MOUSE
|
||||||
| NC score | 0.005741 (rank : 185) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.005226 (rank : 186) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
NETR_MOUSE
|
||||||
| NC score | 0.004982 (rank : 187) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08762 | Gene names | Prss12, Bssp3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Brain-specific serine protease 3) (BSSP-3). | |||||
|
CXA12_MOUSE
|
||||||
| NC score | 0.004558 (rank : 188) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BQU6, Q6TLV2, Q8BQS2, Q9EPM1 | Gene names | Gja12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gap junction alpha-12 protein (Connexin-47) (Cx47). | |||||
|
HIC2_MOUSE
|
||||||
| NC score | 0.003287 (rank : 189) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JLZ6, Q3U030, Q3ULP4, Q5K036, Q8BSZ9, Q8C3T5 | Gene names | Hic2, Kiaa1020 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 2 protein (Hic-2). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.002682 (rank : 190) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
ZN263_HUMAN
|
||||||
| NC score | 0.001723 (rank : 191) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14978, O43387, Q96H95 | Gene names | ZNF263, FPM315 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 263 (Zinc finger protein FPM315). | |||||
|
ZNF74_HUMAN
|
||||||
| NC score | 0.001244 (rank : 192) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16587, Q6IBV2, Q9UC04, Q9UF05, Q9UF06, Q9UF07 | Gene names | ZNF74 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 74 (hZNF7). | |||||
|
ZN192_HUMAN
|
||||||
| NC score | 0.000460 (rank : 193) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 170 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15776, Q4VAR1, Q4VAR2, Q4VAR3, Q9H4T1 | Gene names | ZNF192 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 192 (LD5-1). | |||||