Please be patient as the page loads
|
PI51C_HUMAN
|
||||||
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PI51A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.982084 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99755, Q99754, Q99756 | Gene names | PIP5K1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I alpha) (PtdIns(4)P- 5-kinase alpha) (PIP5KIalpha) (68 kDa type I phosphatidylinositol-4- phosphate 5-kinase alpha). | |||||
|
PI51A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.983581 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70182, Q8K0D3, Q99L80 | Gene names | Pip5k1a, Pip5k1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I alpha) (PtdIns(4)P- 5-kinase alpha) (PIP5KIalpha) (Phosphatidylinositol-4-phosphate 5- kinase type I beta) (PI4P5KIbeta) (68 kDa type I phosphatidylinositol- 4-phosphate 5-kinase). | |||||
|
PI51C_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
PI51C_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.989471 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O70161, Q505A1, Q80TW9, Q8VCU5 | Gene names | Pip5k1c, Kiaa0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
PI51B_MOUSE
|
||||||
| θ value | 6.11349e-182 (rank : 5) | NC score | 0.981046 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70181, O70335, Q8JZY6 | Gene names | Pip5k1b, Pip5k1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 beta (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I beta) (PtdIns(4)P-5- kinase beta) (PIP5KIbeta) (Phosphatidylinositol-4-phosphate 5-kinase type I alpha) (PIP5KIalpha). | |||||
|
PI51B_HUMAN
|
||||||
| θ value | 2.48782e-175 (rank : 6) | NC score | 0.980909 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14986, P78518, P78519, Q5T5K6, Q5T5K8, Q5T5K9, Q5VZ00, Q7KYT5, Q8NHQ5, Q92749 | Gene names | PIP5K1B, STM7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 beta (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I beta) (PtdIns(4)P-5- kinase beta) (PIP5KIbeta) (Type I phosphatidylinositol-4-phosphate 5- kinase beta) (Protein STM-7). | |||||
|
PI52B_HUMAN
|
||||||
| θ value | 4.568e-36 (rank : 7) | NC score | 0.819453 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P78356, Q5U0E8, Q8TBP2 | Gene names | PIP5K2B | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 beta (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II beta) (1- phosphatidylinositol-4-phosphate 5-kinase 2-beta) (PtdIns(4)P-5-kinase isoform 2-beta) (PIP5KII-beta) (Diphosphoinositide kinase 2-beta). | |||||
|
PI52B_MOUSE
|
||||||
| θ value | 4.568e-36 (rank : 8) | NC score | 0.819799 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80XI4, Q8VHB2 | Gene names | Pip5k2b | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 beta (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II beta) (1- phosphatidylinositol-4-phosphate 5-kinase 2-beta) (PtdIns(4)P-5-kinase isoform 2-beta) (PIP5KII-beta) (Diphosphoinositide kinase 2-beta). | |||||
|
PI52A_HUMAN
|
||||||
| θ value | 1.6247e-33 (rank : 9) | NC score | 0.811583 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48426, P53807, Q5VUX3 | Gene names | PIP5K2A, PIP5K2 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II alpha) (1- phosphatidylinositol-4-phosphate 5-kinase 2-alpha) (PtdIns(4)P-5- kinase isoform 2-alpha) (PIP5KII-alpha) (Diphosphoinositide kinase 2- alpha) (PtdIns(4)P-5-kinase B isoform) (PIP5KIII) (PtdIns(4)P-5-kinase C isoform). | |||||
|
PI52A_MOUSE
|
||||||
| θ value | 1.0531e-32 (rank : 10) | NC score | 0.809647 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O70172 | Gene names | Pip5k2a | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II alpha) (1- phosphatidylinositol-4-phosphate 5-kinase 2-alpha) (PtdIns(4)P-5- kinase isoform 2-alpha) (PIP5KII-alpha) (Diphosphoinositide kinase 2- alpha). | |||||
|
FYV1_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 11) | NC score | 0.325742 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
FYV1_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 12) | NC score | 0.314778 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 13) | NC score | 0.053318 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ACK1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 14) | NC score | 0.007677 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.054002 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.030868 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.049035 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.038244 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
BASP_HUMAN
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.056976 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
PALB2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.031995 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3U0P1, Q6NZG9, Q7TMQ4, Q8CEA9 | Gene names | Palb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.034191 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
RFFL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.027346 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZ73, Q8NHW0, Q8TBY7, Q96BE6 | Gene names | RFFL, RNF189, RNF34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (FYVE-RING finger protein Sakura) (Fring) (Caspases-8 and -10- associated RING finger protein 2) (CARP-2) (Caspase regulator CARP2) (RING finger protein 189) (RING finger protein 34-like). | |||||
|
RFIP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.049931 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.049151 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.035812 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.017496 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.033551 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
TNNT2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.020355 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50752, Q64360, Q64377 | Gene names | Tnnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.053068 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.008871 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.031264 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.047452 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.010795 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.012794 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.024687 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.008977 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.019428 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
ANR41_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.013031 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.003764 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.017228 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.028450 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
PHC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.017381 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
RBM19_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.011827 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R3C6, Q8BHR0, Q9CW63 | Gene names | Rbm19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
RTN2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.022851 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70622, O70620 | Gene names | Rtn2, Nspl1 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-2 (Neuroendocrine-specific protein-like 1) (NSP-like protein 1) (NSPLI). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.021818 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.049311 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
EDRF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.018947 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6GQV7, Q80ZY3, Q8R3P5 | Gene names | Edrf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Erythroid differentiation-related factor 1. | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.027598 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.012573 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
JIP2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.018309 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
LRCH4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.006579 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75427, Q8WV85, Q96ID0 | Gene names | LRCH4, LRN, LRRN4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4 (Leucine-rich neuronal protein). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.037139 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.051085 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
BRD3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.012711 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
HMGA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.049329 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17096, P10910, Q9UKB0 | Gene names | HMGA1, HMGIY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMG-I/HMG-Y (HMG-I(Y)) (High mobility group AT-hook protein 1) (High mobility group protein A1) (High mobility group protein-R). | |||||
|
MAP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.018896 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
NCKX2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.016760 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UI40, Q9NTN5, Q9NZQ4 | Gene names | SLC24A2, NCKX2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 2 precursor (Na(+)/K(+)/Ca(2+)- exchange protein 2) (Retinal cone Na-Ca+K exchanger) (Solute carrier family 24 member 2). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.022318 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.038344 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.007246 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.021834 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.023977 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.017012 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.039054 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
ACK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.004642 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |
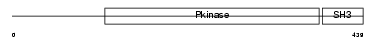
|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
BIR1G_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.015034 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.009309 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DNM3A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.013826 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88508, Q3UH24, Q8CJ60, Q922J0, Q9CSE1 | Gene names | Dnmt3a | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase MmuIIIA) (DNA MTase MmuIIIA) (M.MmuIIIA). | |||||
|
MAGB6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.012050 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.030510 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.012748 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NVL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.008034 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
RGP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.010950 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P47736 | Gene names | RAP1GAP, RAP1GA1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-activating protein 1 (Rap1GAP). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.026626 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
BCLF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.014683 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
BIR1E_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.014569 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
BIR1F_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.014592 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
COFA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.008309 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
FA8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.015558 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.009612 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
HMGA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.035675 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17095, Q91WV2, Q924L7, Q924L8 | Gene names | Hmga1, Hmgi, Hmgiy | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMG-I/HMG-Y (HMG-I(Y)) (High mobility group AT-hook protein 1) (High mobility group protein A1). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.019467 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
SART3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.007568 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15020, Q58F06, Q8IUS1, Q96J95 | Gene names | SART3, KIAA0156, TIP110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (hSART-3) (Tat-interacting protein of 110 kDa) (Tip110). | |||||
|
TNNT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.014301 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
AIPL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.009041 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.006938 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CMTA2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.011349 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
HEXB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.017708 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20060 | Gene names | Hexb | |||
|
Domain Architecture |

|
|||||
| Description | Beta-hexosaminidase beta chain precursor (EC 3.2.1.52) (N-acetyl-beta- glucosaminidase) (Beta-N-acetylhexosaminidase) (Hexosaminidase B). | |||||
|
IC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.004840 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.022564 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.025644 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.010477 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
NBN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.013671 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60934, O60672, Q32NF7, Q53FM6, Q63HR6, Q7LDM2 | Gene names | NBN, NBS, NBS1, P95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1) (Cell cycle regulatory protein p95). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.010538 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.008979 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
ZN202_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | -0.002908 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95125, Q4JG21, Q9H1B9, Q9NSM4 | Gene names | ZNF202 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 202. | |||||
|
ZN326_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.010098 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.010116 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
BRD3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.011619 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.001429 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CHFR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.006681 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96EP1, Q96SL3, Q9NRT4, Q9NT32, Q9NVD5 | Gene names | CHFR | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase CHFR (EC 6.3.2.-) (Checkpoint with forkhead and RING finger domains protein). | |||||
|
DNM3A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.011908 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
MAGD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.010494 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
NALP5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.003517 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NKX28_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.000441 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70584 | Gene names | Nkx2-8, Nkx-2.9, Nkx2-9, Nkx2h | |||
|
Domain Architecture |
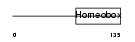
|
|||||
| Description | Homeobox protein Nkx-2.8 (Homeobox protein NK-2 homolog H) (Homeobox protein Nkx-2.9). | |||||
|
PP16A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.002953 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q923M0, Q4V9Z1 | Gene names | Ppp1r16a, Mypt3 | |||
|
Domain Architecture |
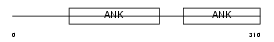
|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase targeting subunit 3). | |||||
|
RP1L1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.022615 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.018952 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TJAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.006927 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DCD5, Q8CFL7, Q8R5I2 | Gene names | Tjap1, Pilt, Tjp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
PI51C_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
PI51C_MOUSE
|
||||||
| NC score | 0.989471 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O70161, Q505A1, Q80TW9, Q8VCU5 | Gene names | Pip5k1c, Kiaa0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
PI51A_MOUSE
|
||||||
| NC score | 0.983581 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70182, Q8K0D3, Q99L80 | Gene names | Pip5k1a, Pip5k1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I alpha) (PtdIns(4)P- 5-kinase alpha) (PIP5KIalpha) (Phosphatidylinositol-4-phosphate 5- kinase type I beta) (PI4P5KIbeta) (68 kDa type I phosphatidylinositol- 4-phosphate 5-kinase). | |||||
|
PI51A_HUMAN
|
||||||
| NC score | 0.982084 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99755, Q99754, Q99756 | Gene names | PIP5K1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I alpha) (PtdIns(4)P- 5-kinase alpha) (PIP5KIalpha) (68 kDa type I phosphatidylinositol-4- phosphate 5-kinase alpha). | |||||
|
PI51B_MOUSE
|
||||||
| NC score | 0.981046 (rank : 5) | θ value | 6.11349e-182 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70181, O70335, Q8JZY6 | Gene names | Pip5k1b, Pip5k1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 beta (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I beta) (PtdIns(4)P-5- kinase beta) (PIP5KIbeta) (Phosphatidylinositol-4-phosphate 5-kinase type I alpha) (PIP5KIalpha). | |||||
|
PI51B_HUMAN
|
||||||
| NC score | 0.980909 (rank : 6) | θ value | 2.48782e-175 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14986, P78518, P78519, Q5T5K6, Q5T5K8, Q5T5K9, Q5VZ00, Q7KYT5, Q8NHQ5, Q92749 | Gene names | PIP5K1B, STM7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 beta (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I beta) (PtdIns(4)P-5- kinase beta) (PIP5KIbeta) (Type I phosphatidylinositol-4-phosphate 5- kinase beta) (Protein STM-7). | |||||
|
PI52B_MOUSE
|
||||||
| NC score | 0.819799 (rank : 7) | θ value | 4.568e-36 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80XI4, Q8VHB2 | Gene names | Pip5k2b | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 beta (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II beta) (1- phosphatidylinositol-4-phosphate 5-kinase 2-beta) (PtdIns(4)P-5-kinase isoform 2-beta) (PIP5KII-beta) (Diphosphoinositide kinase 2-beta). | |||||
|
PI52B_HUMAN
|
||||||
| NC score | 0.819453 (rank : 8) | θ value | 4.568e-36 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P78356, Q5U0E8, Q8TBP2 | Gene names | PIP5K2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 beta (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II beta) (1- phosphatidylinositol-4-phosphate 5-kinase 2-beta) (PtdIns(4)P-5-kinase isoform 2-beta) (PIP5KII-beta) (Diphosphoinositide kinase 2-beta). | |||||
|
PI52A_HUMAN
|
||||||
| NC score | 0.811583 (rank : 9) | θ value | 1.6247e-33 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48426, P53807, Q5VUX3 | Gene names | PIP5K2A, PIP5K2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II alpha) (1- phosphatidylinositol-4-phosphate 5-kinase 2-alpha) (PtdIns(4)P-5- kinase isoform 2-alpha) (PIP5KII-alpha) (Diphosphoinositide kinase 2- alpha) (PtdIns(4)P-5-kinase B isoform) (PIP5KIII) (PtdIns(4)P-5-kinase C isoform). | |||||
|
PI52A_MOUSE
|
||||||
| NC score | 0.809647 (rank : 10) | θ value | 1.0531e-32 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O70172 | Gene names | Pip5k2a | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II alpha) (1- phosphatidylinositol-4-phosphate 5-kinase 2-alpha) (PtdIns(4)P-5- kinase isoform 2-alpha) (PIP5KII-alpha) (Diphosphoinositide kinase 2- alpha). | |||||
|
FYV1_MOUSE
|
||||||
| NC score | 0.325742 (rank : 11) | θ value | 1.38499e-08 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
FYV1_HUMAN
|
||||||
| NC score | 0.314778 (rank : 12) | θ value | 6.87365e-08 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.056976 (rank : 13) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.054002 (rank : 14) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.053318 (rank : 15) | θ value | 0.0148317 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.053068 (rank : 16) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.051085 (rank : 17) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
RFIP1_HUMAN
|
||||||
| NC score | 0.049931 (rank : 18) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
HMGA1_HUMAN
|
||||||
| NC score | 0.049329 (rank : 19) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17096, P10910, Q9UKB0 | Gene names | HMGA1, HMGIY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMG-I/HMG-Y (HMG-I(Y)) (High mobility group AT-hook protein 1) (High mobility group protein A1) (High mobility group protein-R). | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.049311 (rank : 20) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.049151 (rank : 21) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.049035 (rank : 22) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.047452 (rank : 23) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.039054 (rank : 24) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.038344 (rank : 25) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.038244 (rank : 26) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.037139 (rank : 27) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.035812 (rank : 28) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
HMGA1_MOUSE
|
||||||
| NC score | 0.035675 (rank : 29) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17095, Q91WV2, Q924L7, Q924L8 | Gene names | Hmga1, Hmgi, Hmgiy | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMG-I/HMG-Y (HMG-I(Y)) (High mobility group AT-hook protein 1) (High mobility group protein A1). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.034191 (rank : 30) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.033551 (rank : 31) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
PALB2_MOUSE
|
||||||
| NC score | 0.031995 (rank : 32) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3U0P1, Q6NZG9, Q7TMQ4, Q8CEA9 | Gene names | Palb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.031264 (rank : 33) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.030868 (rank : 34) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.030510 (rank : 35) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.028450 (rank : 36) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.027598 (rank : 37) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
RFFL_HUMAN
|
||||||
| NC score | 0.027346 (rank : 38) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZ73, Q8NHW0, Q8TBY7, Q96BE6 | Gene names | RFFL, RNF189, RNF34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (FYVE-RING finger protein Sakura) (Fring) (Caspases-8 and -10- associated RING finger protein 2) (CARP-2) (Caspase regulator CARP2) (RING finger protein 189) (RING finger protein 34-like). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.026626 (rank : 39) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.025644 (rank : 40) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.024687 (rank : 41) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.023977 (rank : 42) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
RTN2_MOUSE
|
||||||
| NC score | 0.022851 (rank : 43) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70622, O70620 | Gene names | Rtn2, Nspl1 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-2 (Neuroendocrine-specific protein-like 1) (NSP-like protein 1) (NSPLI). | |||||
|
RP1L1_MOUSE
|
||||||
| NC score | 0.022615 (rank : 44) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.022564 (rank : 45) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.022318 (rank : 46) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.021834 (rank : 47) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.021818 (rank : 48) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TNNT2_MOUSE
|
||||||
| NC score | 0.020355 (rank : 49) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50752, Q64360, Q64377 | Gene names | Tnnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.019467 (rank : 50) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.019428 (rank : 51) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.018952 (rank : 52) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
EDRF1_MOUSE
|
||||||
| NC score | 0.018947 (rank : 53) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6GQV7, Q80ZY3, Q8R3P5 | Gene names | Edrf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Erythroid differentiation-related factor 1. | |||||
|
MAP2_HUMAN
|
||||||
| NC score | 0.018896 (rank : 54) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
JIP2_MOUSE
|
||||||
| NC score | 0.018309 (rank : 55) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
HEXB_MOUSE
|
||||||
| NC score | 0.017708 (rank : 56) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20060 | Gene names | Hexb | |||
|
Domain Architecture |

|
|||||
| Description | Beta-hexosaminidase beta chain precursor (EC 3.2.1.52) (N-acetyl-beta- glucosaminidase) (Beta-N-acetylhexosaminidase) (Hexosaminidase B). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.017496 (rank : 57) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
PHC2_HUMAN
|
||||||
| NC score | 0.017381 (rank : 58) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.017228 (rank : 59) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.017012 (rank : 60) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
NCKX2_HUMAN
|
||||||
| NC score | 0.016760 (rank : 61) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UI40, Q9NTN5, Q9NZQ4 | Gene names | SLC24A2, NCKX2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 2 precursor (Na(+)/K(+)/Ca(2+)- exchange protein 2) (Retinal cone Na-Ca+K exchanger) (Solute carrier family 24 member 2). | |||||
|
FA8_HUMAN
|
||||||
| NC score | 0.015558 (rank : 62) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
BIR1G_MOUSE
|
||||||
| NC score | 0.015034 (rank : 63) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
BCLF1_HUMAN
|
||||||
| NC score | 0.014683 (rank : 64) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
BIR1F_MOUSE
|
||||||
| NC score | 0.014592 (rank : 65) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1E_MOUSE
|
||||||
| NC score | 0.014569 (rank : 66) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
TNNT2_HUMAN
|
||||||
| NC score | 0.014301 (rank : 67) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
DNM3A_MOUSE
|
||||||
| NC score | 0.013826 (rank : 68) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88508, Q3UH24, Q8CJ60, Q922J0, Q9CSE1 | Gene names | Dnmt3a | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase MmuIIIA) (DNA MTase MmuIIIA) (M.MmuIIIA). | |||||
|
NBN_HUMAN
|
||||||
| NC score | 0.013671 (rank : 69) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60934, O60672, Q32NF7, Q53FM6, Q63HR6, Q7LDM2 | Gene names | NBN, NBS, NBS1, P95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1) (Cell cycle regulatory protein p95). | |||||
|
ANR41_HUMAN
|
||||||
| NC score | 0.013031 (rank : 70) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.012794 (rank : 71) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.012748 (rank : 72) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BRD3_HUMAN
|
||||||
| NC score | 0.012711 (rank : 73) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.012573 (rank : 74) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
MAGB6_HUMAN
|
||||||
| NC score | 0.012050 (rank : 75) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
DNM3A_HUMAN
|
||||||
| NC score | 0.011908 (rank : 76) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
RBM19_MOUSE
|
||||||
| NC score | 0.011827 (rank : 77) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R3C6, Q8BHR0, Q9CW63 | Gene names | Rbm19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
BRD3_MOUSE
|
||||||
| NC score | 0.011619 (rank : 78) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
CMTA2_MOUSE
|
||||||
| NC score | 0.011349 (rank : 79) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
RGP2_HUMAN
|
||||||
| NC score | 0.010950 (rank : 80) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P47736 | Gene names | RAP1GAP, RAP1GA1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-activating protein 1 (Rap1GAP). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.010795 (rank : 81) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.010538 (rank : 82) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
MAGD1_HUMAN
|
||||||
| NC score | 0.010494 (rank : 83) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.010477 (rank : 84) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.010116 (rank : 85) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
ZN326_MOUSE
|
||||||
| NC score | 0.010098 (rank : 86) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.009612 (rank : 87) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.009309 (rank : 88) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
AIPL1_HUMAN
|
||||||
| NC score | 0.009041 (rank : 89) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.008979 (rank : 90) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.008977 (rank : 91) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.008871 (rank : 92) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
COFA1_HUMAN
|
||||||
| NC score | 0.008309 (rank : 93) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
NVL_HUMAN
|
||||||
| NC score | 0.008034 (rank : 94) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
ACK1_HUMAN
|
||||||
| NC score | 0.007677 (rank : 95) | θ value | 0.0431538 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
SART3_HUMAN
|
||||||
| NC score | 0.007568 (rank : 96) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15020, Q58F06, Q8IUS1, Q96J95 | Gene names | SART3, KIAA0156, TIP110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (hSART-3) (Tat-interacting protein of 110 kDa) (Tip110). | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.007246 (rank : 97) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.006938 (rank : 98) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
TJAP1_MOUSE
|
||||||
| NC score | 0.006927 (rank : 99) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DCD5, Q8CFL7, Q8R5I2 | Gene names | Tjap1, Pilt, Tjp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
CHFR_HUMAN
|
||||||
| NC score | 0.006681 (rank : 100) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96EP1, Q96SL3, Q9NRT4, Q9NT32, Q9NVD5 | Gene names | CHFR | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase CHFR (EC 6.3.2.-) (Checkpoint with forkhead and RING finger domains protein). | |||||
|
LRCH4_HUMAN
|
||||||
| NC score | 0.006579 (rank : 101) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75427, Q8WV85, Q96ID0 | Gene names | LRCH4, LRN, LRRN4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4 (Leucine-rich neuronal protein). | |||||
|
IC1_MOUSE
|
||||||
| NC score | 0.004840 (rank : 102) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
ACK1_MOUSE
|
||||||
| NC score | 0.004642 (rank : 103) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | 0.003764 (rank : 104) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
NALP5_HUMAN
|
||||||
| NC score | 0.003517 (rank : 105) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
PP16A_MOUSE
|
||||||
| NC score | 0.002953 (rank : 106) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q923M0, Q4V9Z1 | Gene names | Ppp1r16a, Mypt3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase targeting subunit 3). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.001429 (rank : 107) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
NKX28_MOUSE
|
||||||
| NC score | 0.000441 (rank : 108) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70584 | Gene names | Nkx2-8, Nkx-2.9, Nkx2-9, Nkx2h | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.8 (Homeobox protein NK-2 homolog H) (Homeobox protein Nkx-2.9). | |||||
|
ZN202_HUMAN
|
||||||
| NC score | -0.002908 (rank : 109) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95125, Q4JG21, Q9H1B9, Q9NSM4 | Gene names | ZNF202 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 202. | |||||