Please be patient as the page loads
|
FA8_HUMAN
|
||||||
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FA8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
FA8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989977 (rank : 2) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
FA5_HUMAN
|
||||||
| θ value | 1.46868e-151 (rank : 3) | NC score | 0.931033 (rank : 3) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
HEPH_MOUSE
|
||||||
| θ value | 5.8157e-124 (rank : 4) | NC score | 0.698293 (rank : 7) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0Z4, Q6ZQ65, Q80Y80, Q8C4S2 | Gene names | Heph, Kiaa0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
CERU_HUMAN
|
||||||
| θ value | 2.52853e-119 (rank : 5) | NC score | 0.696921 (rank : 8) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P00450, Q14063, Q2PP18, Q9UKS4 | Gene names | CP | |||
|
Domain Architecture |
|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
HEPH_HUMAN
|
||||||
| θ value | 2.00348e-116 (rank : 6) | NC score | 0.695653 (rank : 9) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQS7, O75180, Q6UW45, Q9C058 | Gene names | HEPH, KIAA0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
CERU_MOUSE
|
||||||
| θ value | 1.02895e-112 (rank : 7) | NC score | 0.694526 (rank : 10) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61147 | Gene names | Cp | |||
|
Domain Architecture |

|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
MFGM_MOUSE
|
||||||
| θ value | 5.91091e-68 (rank : 8) | NC score | 0.666429 (rank : 11) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |
|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
EDIL3_HUMAN
|
||||||
| θ value | 1.00825e-67 (rank : 9) | NC score | 0.550544 (rank : 15) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EDIL3_MOUSE
|
||||||
| θ value | 6.53529e-67 (rank : 10) | NC score | 0.541861 (rank : 16) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
MFGM_HUMAN
|
||||||
| θ value | 8.55514e-59 (rank : 11) | NC score | 0.766309 (rank : 4) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q08431 | Gene names | MFGE8 | |||
|
Domain Architecture |
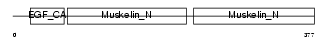
|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin]. | |||||
|
NRP1_MOUSE
|
||||||
| θ value | 3.61106e-41 (rank : 12) | NC score | 0.533166 (rank : 19) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
NRP2_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 13) | NC score | 0.537143 (rank : 18) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP2_MOUSE
|
||||||
| θ value | 3.99248e-40 (rank : 14) | NC score | 0.539580 (rank : 17) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP1_HUMAN
|
||||||
| θ value | 3.37985e-39 (rank : 15) | NC score | 0.530262 (rank : 20) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
DCBD2_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 16) | NC score | 0.576457 (rank : 14) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
DCBD2_MOUSE
|
||||||
| θ value | 2.12685e-25 (rank : 17) | NC score | 0.588061 (rank : 12) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |
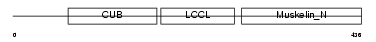
|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
DCBD1_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 18) | NC score | 0.588013 (rank : 13) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
CPXM2_MOUSE
|
||||||
| θ value | 4.9032e-22 (rank : 19) | NC score | 0.481624 (rank : 21) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D2L5, O54860, Q8VDQ4 | Gene names | Cpxm2, Cpx2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CPXM2_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 20) | NC score | 0.478823 (rank : 22) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N436 | Gene names | CPXM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
XLRS1_MOUSE
|
||||||
| θ value | 3.51386e-20 (rank : 21) | NC score | 0.729240 (rank : 5) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z1L4 | Gene names | Rs1, Rs1h, Xlrs1 | |||
|
Domain Architecture |
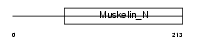
|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein homolog). | |||||
|
XLRS1_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 22) | NC score | 0.728464 (rank : 6) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15537 | Gene names | RS1, XLRS1 | |||
|
Domain Architecture |
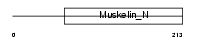
|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein). | |||||
|
CNTP2_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 23) | NC score | 0.463066 (rank : 23) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UHC6, Q52LV1, Q5H9Q7, Q9UQ12 | Gene names | CNTNAP2, CASPR2, KIAA0868 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CPXM1_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 24) | NC score | 0.434363 (rank : 29) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z100, Q99LA3 | Gene names | Cpxm1, Cpx1, Cpxm | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CPXM1_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 25) | NC score | 0.442074 (rank : 28) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96SM3, Q6P4G8, Q6UW65, Q9NUB5 | Gene names | CPXM1, CPX1, CPXM | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CNTP4_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 26) | NC score | 0.446204 (rank : 26) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99P47 | Gene names | Cntnap4, Caspr4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP4_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 27) | NC score | 0.443397 (rank : 27) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP2_MOUSE
|
||||||
| θ value | 7.58209e-15 (rank : 28) | NC score | 0.450711 (rank : 25) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CPW0, Q6P2K4, Q6ZQ31 | Gene names | Cntnap2, Kiaa0868 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP3_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 29) | NC score | 0.453587 (rank : 24) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
DDR2_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 30) | NC score | 0.164178 (rank : 32) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62371 | Gene names | Ddr2, Ntrkr3, Tkt, Tyro10 | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin domain-containing receptor 2 precursor (EC 2.7.10.1) (Discoidin domain receptor 2) (Receptor protein-tyrosine kinase TKT) (Tyrosine-protein kinase TYRO 10) (Neurotrophic tyrosine kinase, receptor-related 3) (CD167b antigen). | |||||
|
DDR1_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 31) | NC score | 0.150869 (rank : 35) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q08345, Q14196, Q16562, Q5ST12, Q9UD37 | Gene names | DDR1, CAK, EDDR1, RTK6, TRKE | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial discoidin domain-containing receptor 1 precursor (EC 2.7.10.1) (Epithelial discoidin domain receptor 1) (Tyrosine kinase DDR) (Discoidin receptor tyrosine kinase) (Tyrosine-protein kinase CAK) (Cell adhesion kinase) (TRK E) (Protein-tyrosine kinase RTK 6) (HGK2) (CD167a antigen). | |||||
|
CNTP1_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 32) | NC score | 0.421300 (rank : 30) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
DDR1_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 33) | NC score | 0.143975 (rank : 36) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q03146 | Gene names | Ddr1, Cak, Eddr1, Mpk6 | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial discoidin domain-containing receptor 1 precursor (EC 2.7.10.1) (Epithelial discoidin domain receptor 1) (Tyrosine kinase DDR) (Discoidin receptor tyrosine kinase) (Tyrosine-protein kinase CAK) (Cell adhesion kinase) (Protein-tyrosine kinase MPK-6) (CD167a antigen). | |||||
|
DDR2_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 34) | NC score | 0.162424 (rank : 33) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16832 | Gene names | DDR2, NTRKR3, TKT, TYRO10 | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin domain-containing receptor 2 precursor (EC 2.7.10.1) (Discoidin domain receptor 2) (Receptor protein-tyrosine kinase TKT) (Tyrosine-protein kinase TYRO 10) (Neurotrophic tyrosine kinase, receptor-related 3) (CD167b antigen). | |||||
|
CNTP1_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 35) | NC score | 0.419184 (rank : 31) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O54991 | Gene names | Cntnap1, Nrxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (Paranodin) (NCP1) (MHDNIV). | |||||
|
CALB2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 36) | NC score | 0.047764 (rank : 95) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 37) | NC score | 0.017174 (rank : 118) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 38) | NC score | 0.017167 (rank : 119) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
ZN281_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 39) | NC score | 0.002639 (rank : 160) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2X9, Q9NY92 | Gene names | ZNF281, GZP1, ZBP99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 281 (Zinc finger DNA-binding protein 99) (Transcription factor ZBP-99) (GC-box-binding zinc finger protein 1). | |||||
|
CALB2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 40) | NC score | 0.046698 (rank : 97) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
TM117_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 41) | NC score | 0.056292 (rank : 83) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BH18, Q8C6X8 | Gene names | Tmem117 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 117. | |||||
|
BORG2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 42) | NC score | 0.050387 (rank : 93) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQC5, Q3UD77, Q8BVR7 | Gene names | Cdc42ep3, Borg2, Cep3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 3 (Binder of Rho GTPases 2). | |||||
|
BORG2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 43) | NC score | 0.049593 (rank : 94) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKI2, O95353, Q9UQJ0 | Gene names | CDC42EP3, BORG2, CEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 3 (Binder of Rho GTPases 2) (MSE55-related Cdc42-binding protein). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 44) | NC score | 0.047021 (rank : 96) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
K0319_MOUSE
|
||||||
| θ value | 0.163984 (rank : 45) | NC score | 0.028602 (rank : 99) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SZV5, Q80U39 | Gene names | Kiaa0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
TOP2B_HUMAN
|
||||||
| θ value | 0.163984 (rank : 46) | NC score | 0.020983 (rank : 107) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02880, Q13600, Q9UMG8, Q9UQP8 | Gene names | TOP2B | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-beta (EC 5.99.1.3) (DNA topoisomerase II, beta isozyme). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 47) | NC score | 0.021624 (rank : 104) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
RTN4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 48) | NC score | 0.022523 (rank : 103) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NQC3, O94962, Q9BXG5, Q9H212, Q9H3I3, Q9UQ42, Q9Y293, Q9Y2Y7, Q9Y5U6 | Gene names | RTN4, KIAA0886, NOGO | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.015000 (rank : 125) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.025709 (rank : 101) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
TTP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.021404 (rank : 105) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
VASP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.018074 (rank : 115) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
KI21B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.008302 (rank : 139) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
ZN286_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | -0.002350 (rank : 167) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBT8, Q96JF3 | Gene names | ZNF286, KIAA1874 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 286. | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.019802 (rank : 109) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
HIRA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.009093 (rank : 138) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54198, Q8IXN2 | Gene names | HIRA, DGCR1, HIR, TUPLE1 | |||
|
Domain Architecture |
|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
KI21B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.008097 (rank : 140) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
LNX1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.007218 (rank : 143) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TBB1, Q8N4C2, Q96MJ7, Q9BY20 | Gene names | LNX1, LNX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase LNX (EC 6.3.2.-) (Numb-binding protein 1) (Ligand of Numb-protein X 1). | |||||
|
TERF2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.028356 (rank : 100) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15554 | Gene names | TERF2, TRF2 | |||
|
Domain Architecture |
|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
TM117_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.039075 (rank : 98) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0C3 | Gene names | TMEM117 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 117. | |||||
|
LEUK_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.019064 (rank : 113) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
NIBA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.019187 (rank : 112) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
CP17A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.003573 (rank : 156) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05093, Q5TZV7 | Gene names | CYP17A1, CYP17, S17AH | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 17A1 (EC 1.14.99.9) (CYPXVII) (P450-C17) (P450c17) (Steroid 17-alpha-monooxygenase) (Steroid 17-alpha-hydroxylase/17,20 lyase). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.006774 (rank : 144) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TOM70_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.021092 (rank : 106) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.010269 (rank : 132) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
PH4H_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.009360 (rank : 137) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00439, Q16717, Q8TC14 | Gene names | PAH | |||
|
Domain Architecture |
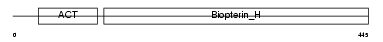
|
|||||
| Description | Phenylalanine-4-hydroxylase (EC 1.14.16.1) (PAH) (Phe-4- monooxygenase). | |||||
|
PI51A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.017780 (rank : 116) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70182, Q8K0D3, Q99L80 | Gene names | Pip5k1a, Pip5k1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I alpha) (PtdIns(4)P- 5-kinase alpha) (PIP5KIalpha) (Phosphatidylinositol-4-phosphate 5- kinase type I beta) (PI4P5KIbeta) (68 kDa type I phosphatidylinositol- 4-phosphate 5-kinase). | |||||
|
TDRD6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.016118 (rank : 122) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
TOM70_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.019798 (rank : 110) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
C2TA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.016582 (rank : 121) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
PARP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.012746 (rank : 127) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
PI51A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.017204 (rank : 117) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99755, Q99754, Q99756 | Gene names | PIP5K1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I alpha) (PtdIns(4)P- 5-kinase alpha) (PIP5KIalpha) (68 kDa type I phosphatidylinositol-4- phosphate 5-kinase alpha). | |||||
|
PKN3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.003351 (rank : 157) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.011378 (rank : 129) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
RGPD8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.020396 (rank : 108) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.003629 (rank : 155) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
AF10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.010849 (rank : 131) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AL2S2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.000028 (rank : 164) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K4T3, Q6P922, Q8BU86, Q8BXK0, Q8K4T2 | Gene names | Als2cr2, Papk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ALS2CR2 (EC 2.7.11.1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 2 protein homolog) (ILP-interacting protein homolog) (Polyploidy-associated protein kinase). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.004500 (rank : 151) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CMTA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.009900 (rank : 135) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6Y1, Q5VUE1 | Gene names | CAMTA1, KIAA0833 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 1. | |||||
|
K0082_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.018979 (rank : 114) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.005042 (rank : 149) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
PI51C_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.016595 (rank : 120) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O70161, Q505A1, Q80TW9, Q8VCU5 | Gene names | Pip5k1c, Kiaa0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
RBP22_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.019781 (rank : 111) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
SOX4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.004480 (rank : 152) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.010895 (rank : 130) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
ZN485_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | -0.003048 (rank : 168) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NCK3, Q96CL0 | Gene names | ZNF485 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 485. | |||||
|
AL2S2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | -0.000626 (rank : 166) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9C0K7, Q5BKY7, Q9P1L0 | Gene names | ALS2CR2, ILPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ALS2CR2 (EC 2.7.11.1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 2 protein) (ILP- interacting protein) (CALS-21). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.010009 (rank : 133) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.015024 (rank : 124) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.024675 (rank : 102) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.005756 (rank : 148) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.006640 (rank : 145) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
PI51C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.015558 (rank : 123) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
TM45A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.013409 (rank : 126) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NWC5, Q53YW5 | Gene names | TMEM45A, DERP7, DNAPTP4 | |||
|
Domain Architecture |
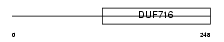
|
|||||
| Description | Transmembrane protein 45a (Dermal papilla-derived protein 7) (DNA polymerase-transactivated protein 4). | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.002749 (rank : 159) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
CI093_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.002943 (rank : 158) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
GP73_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.007247 (rank : 142) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91XA2 | Gene names | Golph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
KIF9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.006450 (rank : 146) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HAQ2, Q86Z28, Q9H8A4 | Gene names | KIF9 | |||
|
Domain Architecture |
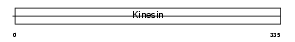
|
|||||
| Description | Kinesin-like protein KIF9. | |||||
|
PCDH9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | -0.000139 (rank : 165) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HC56 | Gene names | PCDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-9 precursor. | |||||
|
RERE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.003683 (rank : 154) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
ASAH2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.007375 (rank : 141) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHE3, Q3UWP9, Q8BNP0, Q8BQN7, Q8R236 | Gene names | Asah2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neutral ceramidase (EC 3.5.1.23) (NCDase) (N-CDase) (Acylsphingosine deacylase 2) (N-acylsphingosine amidohydrolase 2) [Contains: Neutral ceramidase soluble form]. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.002212 (rank : 161) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.003716 (rank : 153) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.001305 (rank : 162) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
KIF5A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.004582 (rank : 150) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
LAS1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.009911 (rank : 134) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4W2, Q5JXQ0, Q8TEN5, Q9H9V5 | Gene names | LAS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LAS1-like protein. | |||||
|
TARA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.009876 (rank : 136) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TLR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.000951 (rank : 163) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15399, O15452, Q9UG90 | Gene names | TLR1, KIAA0012 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 1 precursor (Toll/interleukin-1 receptor-like protein) (TIL) (CD281 antigen). | |||||
|
UT14A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.006132 (rank : 147) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BVJ6, Q5JYF1 | Gene names | UTP14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Antigen NY-CO- 16). | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.011605 (rank : 128) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.064335 (rank : 78) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
BMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.075743 (rank : 69) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
BMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.075624 (rank : 70) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
CBPD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.086468 (rank : 63) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75976, O15377, Q86SH9, Q86XE6 | Gene names | CPD | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CBPD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.086458 (rank : 64) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O89001 | Gene names | Cpd | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CBPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.100323 (rank : 43) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16870, Q9UIU9 | Gene names | CPE | |||
|
Domain Architecture |
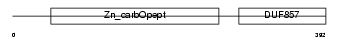
|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.100209 (rank : 44) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00493, Q64439 | Gene names | Cpe | |||
|
Domain Architecture |
|
|||||
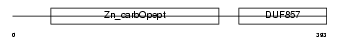
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.089862 (rank : 55) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14384, Q9H2K9 | Gene names | CPM | |||
|
Domain Architecture |
|
|||||
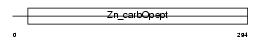
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
CBPM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.090208 (rank : 53) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80V42, Q497S5, Q9CYH8 | Gene names | Cpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
CBPN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.098632 (rank : 47) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15169 | Gene names | CPN1, ACBP | |||
|
Domain Architecture |
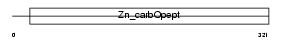
|
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit) (Lysine carboxypeptidase) (Arginine carboxypeptidase) (Kininase-1) (Serum carboxypeptidase N) (SCPN) (Anaphylatoxin inactivator) (Plasma carboxypeptidase B). | |||||
|
CBPN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.098972 (rank : 46) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJN5, Q91WM9 | Gene names | Cpn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit). | |||||
|
CBPZ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.088082 (rank : 58) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q66K79, O00520, Q96MX2 | Gene names | CPZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
CBPZ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.089420 (rank : 56) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R4V4 | Gene names | Cpz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
CRUM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.051446 (rank : 92) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CRUM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.052172 (rank : 91) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CSMD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.052526 (rank : 88) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CSMD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.052187 (rank : 90) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.056254 (rank : 84) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
CSMD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.053116 (rank : 87) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
CUBN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.086775 (rank : 62) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
CUBN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.087241 (rank : 61) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
CUZD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.087552 (rank : 60) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUZD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.082150 (rank : 65) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
DCBD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.158435 (rank : 34) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DMBT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.064874 (rank : 77) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
GLPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.100918 (rank : 42) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04921, Q92642 | Gene names | GYPC, GPC | |||
|
Domain Architecture |
|
|||||
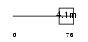
| Description | Glycophorin C (PAS-2') (Glycoprotein beta) (GLPC) (Glycoconnectin) (Sialoglycoprotein D) (Glycophorin D) (GPD) (CD236 antigen). | |||||
|
GP126_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.054747 (rank : 85) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
LRP12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.056324 (rank : 82) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |
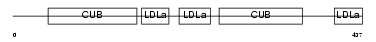
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
LRP12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.058306 (rank : 81) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |
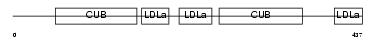
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
MAMC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.068599 (rank : 76) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z304, Q5VW47, Q8WX43, Q96BM4 | Gene names | MAMDC2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 2 precursor. | |||||
|
MAMC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.063995 (rank : 79) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG85, Q8BM24 | Gene names | Mamdc2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 2 precursor. | |||||
|
MFRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.079899 (rank : 67) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MFRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.081050 (rank : 66) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
NETO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.095480 (rank : 51) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.098082 (rank : 48) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.101434 (rank : 41) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NETO2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.099441 (rank : 45) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NRX1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.091062 (rank : 52) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULB1, O60323, Q53TJ9, Q53TQ1, Q9C079, Q9C080, Q9C081, Q9H3M2, Q9UDM6 | Gene names | NRXN1, KIAA0578 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
NRX1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.090014 (rank : 54) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CS84, O88722, Q80Y87, Q8CHE6 | Gene names | Nrxn1, Kiaa0578 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.077844 (rank : 68) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.088036 (rank : 59) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y4C0, O95378, Q9NS47, Q9P1V3, Q9P1V6, Q9UIE2, Q9UIE3, Q9ULA5, Q9Y486 | Gene names | NRXN3, KIAA0743 | |||
|
Domain Architecture |
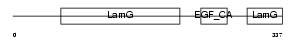
|
|||||
| Description | Neurexin-3-alpha precursor (Neurexin III-alpha). | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.112312 (rank : 39) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
PCOC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.114100 (rank : 38) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
PCOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.111431 (rank : 40) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PCOC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.114209 (rank : 37) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PDGFD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.088568 (rank : 57) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9GZP0, Q9BWV5 | Gene names | PDGFD, IEGF, SCDGFB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor D precursor (PDGF D) (Iris-expressed growth factor) (Spinal cord-derived growth factor B) (SCDGF-B). | |||||
|
PDGFD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.074785 (rank : 74) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q925I7, Q3URF6, Q8K2L3, Q9D1L8 | Gene names | Pdgfd, Scdgfb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor chain precursor (PDGF D) (Spinal cord- derived growth factor B) (SCDGF-B). | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.061183 (rank : 80) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
SHBG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.053378 (rank : 86) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P04278, P14689, Q16616 | Gene names | SHBG | |||
|
Domain Architecture |
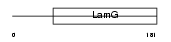
|
|||||
| Description | Sex hormone-binding globulin precursor (SHBG) (Sex steroid-binding protein) (SBP) (Testis-specific androgen-binding protein) (ABP) (Testosterone-estrogen-binding globulin) (Testosterone-estradiol- binding globulin) (TeBG). | |||||
|
TLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.074813 (rank : 72) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
TLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.073956 (rank : 75) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
TLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.074799 (rank : 73) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TLL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.075324 (rank : 71) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TSG6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.096148 (rank : 50) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TSG6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.096649 (rank : 49) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.052497 (rank : 89) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
FA8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
FA8_MOUSE
|
||||||
| NC score | 0.989977 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
FA5_HUMAN
|
||||||
| NC score | 0.931033 (rank : 3) | θ value | 1.46868e-151 (rank : 3) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
MFGM_HUMAN
|
||||||
| NC score | 0.766309 (rank : 4) | θ value | 8.55514e-59 (rank : 11) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q08431 | Gene names | MFGE8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin]. | |||||
|
XLRS1_MOUSE
|
||||||
| NC score | 0.729240 (rank : 5) | θ value | 3.51386e-20 (rank : 21) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z1L4 | Gene names | Rs1, Rs1h, Xlrs1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein homolog). | |||||
|
XLRS1_HUMAN
|
||||||
| NC score | 0.728464 (rank : 6) | θ value | 1.02238e-19 (rank : 22) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15537 | Gene names | RS1, XLRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein). | |||||
|
HEPH_MOUSE
|
||||||
| NC score | 0.698293 (rank : 7) | θ value | 5.8157e-124 (rank : 4) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0Z4, Q6ZQ65, Q80Y80, Q8C4S2 | Gene names | Heph, Kiaa0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
CERU_HUMAN
|
||||||
| NC score | 0.696921 (rank : 8) | θ value | 2.52853e-119 (rank : 5) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P00450, Q14063, Q2PP18, Q9UKS4 | Gene names | CP | |||
|
Domain Architecture |
|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
HEPH_HUMAN
|
||||||
| NC score | 0.695653 (rank : 9) | θ value | 2.00348e-116 (rank : 6) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQS7, O75180, Q6UW45, Q9C058 | Gene names | HEPH, KIAA0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
CERU_MOUSE
|
||||||
| NC score | 0.694526 (rank : 10) | θ value | 1.02895e-112 (rank : 7) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61147 | Gene names | Cp | |||
|
Domain Architecture |

|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
MFGM_MOUSE
|
||||||
| NC score | 0.666429 (rank : 11) | θ value | 5.91091e-68 (rank : 8) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
DCBD2_MOUSE
|
||||||
| NC score | 0.588061 (rank : 12) | θ value | 2.12685e-25 (rank : 17) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
DCBD1_HUMAN
|
||||||
| NC score | 0.588013 (rank : 13) | θ value | 4.01107e-24 (rank : 18) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DCBD2_HUMAN
|
||||||
| NC score | 0.576457 (rank : 14) | θ value | 3.87602e-27 (rank : 16) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
EDIL3_HUMAN
|
||||||
| NC score | 0.550544 (rank : 15) | θ value | 1.00825e-67 (rank : 9) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EDIL3_MOUSE
|
||||||
| NC score | 0.541861 (rank : 16) | θ value | 6.53529e-67 (rank : 10) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
NRP2_MOUSE
|
||||||
| NC score | 0.539580 (rank : 17) | θ value | 3.99248e-40 (rank : 14) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP2_HUMAN
|
||||||
| NC score | 0.537143 (rank : 18) | θ value | 2.34062e-40 (rank : 13) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP1_MOUSE
|
||||||
| NC score | 0.533166 (rank : 19) | θ value | 3.61106e-41 (rank : 12) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
NRP1_HUMAN
|
||||||
| NC score | 0.530262 (rank : 20) | θ value | 3.37985e-39 (rank : 15) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
CPXM2_MOUSE
|
||||||
| NC score | 0.481624 (rank : 21) | θ value | 4.9032e-22 (rank : 19) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D2L5, O54860, Q8VDQ4 | Gene names | Cpxm2, Cpx2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CPXM2_HUMAN
|
||||||
| NC score | 0.478823 (rank : 22) | θ value | 1.86321e-21 (rank : 20) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N436 | Gene names | CPXM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CNTP2_HUMAN
|
||||||
| NC score | 0.463066 (rank : 23) | θ value | 4.02038e-16 (rank : 23) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UHC6, Q52LV1, Q5H9Q7, Q9UQ12 | Gene names | CNTNAP2, CASPR2, KIAA0868 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP3_HUMAN
|
||||||
| NC score | 0.453587 (rank : 24) | θ value | 1.09485e-13 (rank : 29) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
CNTP2_MOUSE
|
||||||
| NC score | 0.450711 (rank : 25) | θ value | 7.58209e-15 (rank : 28) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CPW0, Q6P2K4, Q6ZQ31 | Gene names | Cntnap2, Kiaa0868 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP4_MOUSE
|
||||||
| NC score | 0.446204 (rank : 26) | θ value | 2.60593e-15 (rank : 26) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99P47 | Gene names | Cntnap4, Caspr4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP4_HUMAN
|
||||||
| NC score | 0.443397 (rank : 27) | θ value | 5.8054e-15 (rank : 27) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CPXM1_HUMAN
|
||||||
| NC score | 0.442074 (rank : 28) | θ value | 1.16975e-15 (rank : 25) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96SM3, Q6P4G8, Q6UW65, Q9NUB5 | Gene names | CPXM1, CPX1, CPXM | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CPXM1_MOUSE
|
||||||
| NC score | 0.434363 (rank : 29) | θ value | 4.02038e-16 (rank : 24) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z100, Q99LA3 | Gene names | Cpxm1, Cpx1, Cpxm | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CNTP1_HUMAN
|
||||||
| NC score | 0.421300 (rank : 30) | θ value | 6.00763e-12 (rank : 32) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
CNTP1_MOUSE
|
||||||
| NC score | 0.419184 (rank : 31) | θ value | 6.64225e-11 (rank : 35) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O54991 | Gene names | Cntnap1, Nrxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (Paranodin) (NCP1) (MHDNIV). | |||||
|
DDR2_MOUSE
|
||||||
| NC score | 0.164178 (rank : 32) | θ value | 2.43908e-13 (rank : 30) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62371 | Gene names | Ddr2, Ntrkr3, Tkt, Tyro10 | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin domain-containing receptor 2 precursor (EC 2.7.10.1) (Discoidin domain receptor 2) (Receptor protein-tyrosine kinase TKT) (Tyrosine-protein kinase TYRO 10) (Neurotrophic tyrosine kinase, receptor-related 3) (CD167b antigen). | |||||
|
DDR2_HUMAN
|
||||||
| NC score | 0.162424 (rank : 33) | θ value | 7.84624e-12 (rank : 34) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16832 | Gene names | DDR2, NTRKR3, TKT, TYRO10 | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin domain-containing receptor 2 precursor (EC 2.7.10.1) (Discoidin domain receptor 2) (Receptor protein-tyrosine kinase TKT) (Tyrosine-protein kinase TYRO 10) (Neurotrophic tyrosine kinase, receptor-related 3) (CD167b antigen). | |||||
|
DCBD1_MOUSE
|
||||||
| NC score | 0.158435 (rank : 34) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DDR1_HUMAN
|
||||||
| NC score | 0.150869 (rank : 35) | θ value | 5.43371e-13 (rank : 31) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q08345, Q14196, Q16562, Q5ST12, Q9UD37 | Gene names | DDR1, CAK, EDDR1, RTK6, TRKE | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial discoidin domain-containing receptor 1 precursor (EC 2.7.10.1) (Epithelial discoidin domain receptor 1) (Tyrosine kinase DDR) (Discoidin receptor tyrosine kinase) (Tyrosine-protein kinase CAK) (Cell adhesion kinase) (TRK E) (Protein-tyrosine kinase RTK 6) (HGK2) (CD167a antigen). | |||||
|
DDR1_MOUSE
|
||||||
| NC score | 0.143975 (rank : 36) | θ value | 6.00763e-12 (rank : 33) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q03146 | Gene names | Ddr1, Cak, Eddr1, Mpk6 | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial discoidin domain-containing receptor 1 precursor (EC 2.7.10.1) (Epithelial discoidin domain receptor 1) (Tyrosine kinase DDR) (Discoidin receptor tyrosine kinase) (Tyrosine-protein kinase CAK) (Cell adhesion kinase) (Protein-tyrosine kinase MPK-6) (CD167a antigen). | |||||
|
PCOC2_MOUSE
|
||||||
| NC score | 0.114209 (rank : 37) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PCOC1_MOUSE
|
||||||
| NC score | 0.114100 (rank : 38) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.112312 (rank : 39) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
PCOC2_HUMAN
|
||||||
| NC score | 0.111431 (rank : 40) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
NETO2_HUMAN
|
||||||
| NC score | 0.101434 (rank : 41) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
GLPC_HUMAN
|
||||||
| NC score | 0.100918 (rank : 42) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04921, Q92642 | Gene names | GYPC, GPC | |||
|
Domain Architecture |

|
|||||
| Description | Glycophorin C (PAS-2') (Glycoprotein beta) (GLPC) (Glycoconnectin) (Sialoglycoprotein D) (Glycophorin D) (GPD) (CD236 antigen). | |||||
|
CBPE_HUMAN
|
||||||
| NC score | 0.100323 (rank : 43) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16870, Q9UIU9 | Gene names | CPE | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
CBPE_MOUSE
|
||||||
| NC score | 0.100209 (rank : 44) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00493, Q64439 | Gene names | Cpe | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase E precursor (EC 3.4.17.10) (CPE) (Carboxypeptidase H) (CPH) (Enkephalin convertase) (Prohormone-processing carboxypeptidase). | |||||
|
NETO2_MOUSE
|
||||||
| NC score | 0.099441 (rank : 45) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
CBPN_MOUSE
|
||||||
| NC score | 0.098972 (rank : 46) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJN5, Q91WM9 | Gene names | Cpn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit). | |||||
|
CBPN_HUMAN
|
||||||
| NC score | 0.098632 (rank : 47) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15169 | Gene names | CPN1, ACBP | |||
|
Domain Architecture |
|
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit) (Lysine carboxypeptidase) (Arginine carboxypeptidase) (Kininase-1) (Serum carboxypeptidase N) (SCPN) (Anaphylatoxin inactivator) (Plasma carboxypeptidase B). | |||||
|
NETO1_MOUSE
|
||||||
| NC score | 0.098082 (rank : 48) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
TSG6_MOUSE
|
||||||
| NC score | 0.096649 (rank : 49) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
TSG6_HUMAN
|
||||||
| NC score | 0.096148 (rank : 50) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
NETO1_HUMAN
|
||||||
| NC score | 0.095480 (rank : 51) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NRX1A_HUMAN
|
||||||
| NC score | 0.091062 (rank : 52) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULB1, O60323, Q53TJ9, Q53TQ1, Q9C079, Q9C080, Q9C081, Q9H3M2, Q9UDM6 | Gene names | NRXN1, KIAA0578 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
CBPM_MOUSE
|
||||||
| NC score | 0.090208 (rank : 53) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80V42, Q497S5, Q9CYH8 | Gene names | Cpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
NRX1A_MOUSE
|
||||||
| NC score | 0.090014 (rank : 54) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CS84, O88722, Q80Y87, Q8CHE6 | Gene names | Nrxn1, Kiaa0578 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
CBPM_HUMAN
|
||||||
| NC score | 0.089862 (rank : 55) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14384, Q9H2K9 | Gene names | CPM | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase M precursor (EC 3.4.17.12) (CPM). | |||||
|
CBPZ_MOUSE
|
||||||
| NC score | 0.089420 (rank : 56) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R4V4 | Gene names | Cpz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
PDGFD_HUMAN
|
||||||
| NC score | 0.088568 (rank : 57) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9GZP0, Q9BWV5 | Gene names | PDGFD, IEGF, SCDGFB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor D precursor (PDGF D) (Iris-expressed growth factor) (Spinal cord-derived growth factor B) (SCDGF-B). | |||||
|
CBPZ_HUMAN
|
||||||
| NC score | 0.088082 (rank : 58) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q66K79, O00520, Q96MX2 | Gene names | CPZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
NRX3A_HUMAN
|
||||||
| NC score | 0.088036 (rank : 59) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y4C0, O95378, Q9NS47, Q9P1V3, Q9P1V6, Q9UIE2, Q9UIE3, Q9ULA5, Q9Y486 | Gene names | NRXN3, KIAA0743 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-3-alpha precursor (Neurexin III-alpha). | |||||
|
CUZD1_HUMAN
|
||||||
| NC score | 0.087552 (rank : 60) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUBN_MOUSE
|
||||||
| NC score | 0.087241 (rank : 61) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
CUBN_HUMAN
|
||||||
| NC score | 0.086775 (rank : 62) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
CBPD_HUMAN
|
||||||
| NC score | 0.086468 (rank : 63) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75976, O15377, Q86SH9, Q86XE6 | Gene names | CPD | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CBPD_MOUSE
|
||||||
| NC score | 0.086458 (rank : 64) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O89001 | Gene names | Cpd | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase D precursor (EC 3.4.17.22) (Metallocarboxypeptidase D) (gp180). | |||||
|
CUZD1_MOUSE
|
||||||
| NC score | 0.082150 (rank : 65) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
MFRP_MOUSE
|
||||||
| NC score | 0.081050 (rank : 66) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MFRP_HUMAN
|
||||||
| NC score | 0.079899 (rank : 67) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.077844 (rank : 68) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
BMP1_HUMAN
|
||||||
| NC score | 0.075743 (rank : 69) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
BMP1_MOUSE
|
||||||
| NC score | 0.075624 (rank : 70) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
TLL2_MOUSE
|
||||||
| NC score | 0.075324 (rank : 71) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TLL1_HUMAN
|
||||||
| NC score | 0.074813 (rank : 72) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
TLL2_HUMAN
|
||||||
| NC score | 0.074799 (rank : 73) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
PDGFD_MOUSE
|
||||||
| NC score | 0.074785 (rank : 74) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q925I7, Q3URF6, Q8K2L3, Q9D1L8 | Gene names | Pdgfd, Scdgfb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor chain precursor (PDGF D) (Spinal cord- derived growth factor B) (SCDGF-B). | |||||
|
TLL1_MOUSE
|
||||||
| NC score | 0.073956 (rank : 75) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
MAMC2_HUMAN
|
||||||
| NC score | 0.068599 (rank : 76) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z304, Q5VW47, Q8WX43, Q96BM4 | Gene names | MAMDC2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 2 precursor. | |||||
|
DMBT1_MOUSE
|
||||||
| NC score | 0.064874 (rank : 77) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.064335 (rank : 78) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
MAMC2_MOUSE
|
||||||
| NC score | 0.063995 (rank : 79) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG85, Q8BM24 | Gene names | Mamdc2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 2 precursor. | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.061183 (rank : 80) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
LRP12_MOUSE
|
||||||
| NC score | 0.058306 (rank : 81) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
LRP12_HUMAN
|
||||||
| NC score | 0.056324 (rank : 82) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
TM117_MOUSE
|
||||||
| NC score | 0.056292 (rank : 83) | θ value | 0.0431538 (rank : 41) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BH18, Q8C6X8 | Gene names | Tmem117 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 117. | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.056254 (rank : 84) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
GP126_HUMAN
|
||||||
| NC score | 0.054747 (rank : 85) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
SHBG_HUMAN
|
||||||
| NC score | 0.053378 (rank : 86) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P04278, P14689, Q16616 | Gene names | SHBG | |||
|
Domain Architecture |

|
|||||
| Description | Sex hormone-binding globulin precursor (SHBG) (Sex steroid-binding protein) (SBP) (Testis-specific androgen-binding protein) (ABP) (Testosterone-estrogen-binding globulin) (Testosterone-estradiol- binding globulin) (TeBG). | |||||
|
CSMD3_HUMAN
|
||||||
| NC score | 0.053116 (rank : 87) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
CSMD1_HUMAN
|
||||||
| NC score | 0.052526 (rank : 88) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.052497 (rank : 89) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CSMD1_MOUSE
|
||||||
| NC score | 0.052187 (rank : 90) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CRUM1_MOUSE
|
||||||
| NC score | 0.052172 (rank : 91) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CRUM1_HUMAN
|
||||||
| NC score | 0.051446 (rank : 92) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
BORG2_MOUSE
|
||||||
| NC score | 0.050387 (rank : 93) | θ value | 0.0563607 (rank : 42) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQC5, Q3UD77, Q8BVR7 | Gene names | Cdc42ep3, Borg2, Cep3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 3 (Binder of Rho GTPases 2). | |||||
|
BORG2_HUMAN
|
||||||
| NC score | 0.049593 (rank : 94) | θ value | 0.0736092 (rank : 43) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKI2, O95353, Q9UQJ0 | Gene names | CDC42EP3, BORG2, CEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 3 (Binder of Rho GTPases 2) (MSE55-related Cdc42-binding protein). | |||||
|
CALB2_MOUSE
|
||||||
| NC score | 0.047764 (rank : 95) | θ value | 0.0330416 (rank : 36) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.047021 (rank : 96) | θ value | 0.0736092 (rank : 44) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
CALB2_HUMAN
|
||||||
| NC score | 0.046698 (rank : 97) | θ value | 0.0431538 (rank : 40) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
TM117_HUMAN
|
||||||
| NC score | 0.039075 (rank : 98) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0C3 | Gene names | TMEM117 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 117. | |||||
|
K0319_MOUSE
|
||||||
| NC score | 0.028602 (rank : 99) | θ value | 0.163984 (rank : 45) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SZV5, Q80U39 | Gene names | Kiaa0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
TERF2_HUMAN
|
||||||
| NC score | 0.028356 (rank : 100) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15554 | Gene names | TERF2, TRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.025709 (rank : 101) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.024675 (rank : 102) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
RTN4_HUMAN
|
||||||
| NC score | 0.022523 (rank : 103) | θ value | 0.21417 (rank : 48) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NQC3, O94962, Q9BXG5, Q9H212, Q9H3I3, Q9UQ42, Q9Y293, Q9Y2Y7, Q9Y5U6 | Gene names | RTN4, KIAA0886, NOGO | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.021624 (rank : 104) | θ value | 0.21417 (rank : 47) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
TTP_MOUSE
|
||||||
| NC score | 0.021404 (rank : 105) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
TOM70_MOUSE
|
||||||
| NC score | 0.021092 (rank : 106) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CZW5 | Gene names | Tomm70a, D16Wsu109e | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
TOP2B_HUMAN
|
||||||
| NC score | 0.020983 (rank : 107) | θ value | 0.163984 (rank : 46) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02880, Q13600, Q9UMG8, Q9UQP8 | Gene names | TOP2B | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-beta (EC 5.99.1.3) (DNA topoisomerase II, beta isozyme). | |||||
|
RGPD8_HUMAN
|
||||||
| NC score | 0.020396 (rank : 108) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.019802 (rank : 109) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
TOM70_HUMAN
|
||||||
| NC score | 0.019798 (rank : 110) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94826 | Gene names | TOMM70A, KIAA0719 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial precursor proteins import receptor (Translocase of outer membrane TOM70). | |||||
|
RBP22_HUMAN
|
||||||
| NC score | 0.019781 (rank : 111) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
NIBA_HUMAN
|
||||||
| NC score | 0.019187 (rank : 112) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
LEUK_HUMAN
|
||||||
| NC score | 0.019064 (rank : 113) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
K0082_HUMAN
|
||||||
| NC score | 0.018979 (rank : 114) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
VASP_HUMAN
|
||||||
| NC score | 0.018074 (rank : 115) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
PI51A_MOUSE
|
||||||
| NC score | 0.017780 (rank : 116) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70182, Q8K0D3, Q99L80 | Gene names | Pip5k1a, Pip5k1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I alpha) (PtdIns(4)P- 5-kinase alpha) (PIP5KIalpha) (Phosphatidylinositol-4-phosphate 5- kinase type I beta) (PI4P5KIbeta) (68 kDa type I phosphatidylinositol- 4-phosphate 5-kinase). | |||||
|
PI51A_HUMAN
|
||||||
| NC score | 0.017204 (rank : 117) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99755, Q99754, Q99756 | Gene names | PIP5K1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I alpha) (PtdIns(4)P- 5-kinase alpha) (PIP5KIalpha) (68 kDa type I phosphatidylinositol-4- phosphate 5-kinase alpha). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.017174 (rank : 118) | θ value | 0.0330416 (rank : 37) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.017167 (rank : 119) | θ value | 0.0330416 (rank : 38) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
PI51C_MOUSE
|
||||||
| NC score | 0.016595 (rank : 120) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O70161, Q505A1, Q80TW9, Q8VCU5 | Gene names | Pip5k1c, Kiaa0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
C2TA_MOUSE
|
||||||
| NC score | 0.016582 (rank : 121) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
TDRD6_MOUSE
|
||||||
| NC score | 0.016118 (rank : 122) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
PI51C_HUMAN
|
||||||
| NC score | 0.015558 (rank : 123) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.015024 (rank : 124) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.015000 (rank : 125) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TM45A_HUMAN
|
||||||
| NC score | 0.013409 (rank : 126) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NWC5, Q53YW5 | Gene names | TMEM45A, DERP7, DNAPTP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protein 45a (Dermal papilla-derived protein 7) (DNA polymerase-transactivated protein 4). | |||||
|
PARP4_HUMAN
|
||||||
| NC score | 0.012746 (rank : 127) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.011605 (rank : 128) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.011378 (rank : 129) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.010895 (rank : 130) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
AF10_MOUSE
|
||||||
| NC score | 0.010849 (rank : 131) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.010269 (rank : 132) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.010009 (rank : 133) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
LAS1L_HUMAN
|
||||||
| NC score | 0.009911 (rank : 134) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4W2, Q5JXQ0, Q8TEN5, Q9H9V5 | Gene names | LAS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LAS1-like protein. | |||||
|
CMTA1_HUMAN
|
||||||
| NC score | 0.009900 (rank : 135) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6Y1, Q5VUE1 | Gene names | CAMTA1, KIAA0833 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 1. | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.009876 (rank : 136) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PH4H_HUMAN
|
||||||
| NC score | 0.009360 (rank : 137) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00439, Q16717, Q8TC14 | Gene names | PAH | |||
|
Domain Architecture |

|
|||||
| Description | Phenylalanine-4-hydroxylase (EC 1.14.16.1) (PAH) (Phe-4- monooxygenase). | |||||
|
HIRA_HUMAN
|
||||||
| NC score | 0.009093 (rank : 138) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54198, Q8IXN2 | Gene names | HIRA, DGCR1, HIR, TUPLE1 | |||
|
Domain Architecture |

|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
KI21B_MOUSE
|
||||||
| NC score | 0.008302 (rank : 139) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
KI21B_HUMAN
|
||||||
| NC score | 0.008097 (rank : 140) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
ASAH2_MOUSE
|
||||||
| NC score | 0.007375 (rank : 141) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHE3, Q3UWP9, Q8BNP0, Q8BQN7, Q8R236 | Gene names | Asah2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neutral ceramidase (EC 3.5.1.23) (NCDase) (N-CDase) (Acylsphingosine deacylase 2) (N-acylsphingosine amidohydrolase 2) [Contains: Neutral ceramidase soluble form]. | |||||
|
GP73_MOUSE
|
||||||
| NC score | 0.007247 (rank : 142) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91XA2 | Gene names | Golph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
LNX1_HUMAN
|
||||||
| NC score | 0.007218 (rank : 143) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TBB1, Q8N4C2, Q96MJ7, Q9BY20 | Gene names | LNX1, LNX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase LNX (EC 6.3.2.-) (Numb-binding protein 1) (Ligand of Numb-protein X 1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.006774 (rank : 144) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.006640 (rank : 145) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
KIF9_HUMAN
|
||||||
| NC score | 0.006450 (rank : 146) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HAQ2, Q86Z28, Q9H8A4 | Gene names | KIF9 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF9. | |||||
|
UT14A_HUMAN
|
||||||
| NC score | 0.006132 (rank : 147) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BVJ6, Q5JYF1 | Gene names | UTP14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Antigen NY-CO- 16). | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.005756 (rank : 148) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.005042 (rank : 149) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
KIF5A_HUMAN
|
||||||
| NC score | 0.004582 (rank : 150) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.004500 (rank : 151) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SOX4_MOUSE
|
||||||
| NC score | 0.004480 (rank : 152) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.003716 (rank : 153) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.003683 (rank : 154) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.003629 (rank : 155) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CP17A_HUMAN
|
||||||
| NC score | 0.003573 (rank : 156) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05093, Q5TZV7 | Gene names | CYP17A1, CYP17, S17AH | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 17A1 (EC 1.14.99.9) (CYPXVII) (P450-C17) (P450c17) (Steroid 17-alpha-monooxygenase) (Steroid 17-alpha-hydroxylase/17,20 lyase). | |||||
|
PKN3_MOUSE
|
||||||
| NC score | 0.003351 (rank : 157) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.002943 (rank : 158) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.002749 (rank : 159) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
ZN281_HUMAN
|
||||||
| NC score | 0.002639 (rank : 160) | θ value | 0.0330416 (rank : 39) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2X9, Q9NY92 | Gene names | ZNF281, GZP1, ZBP99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 281 (Zinc finger DNA-binding protein 99) (Transcription factor ZBP-99) (GC-box-binding zinc finger protein 1). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.002212 (rank : 161) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.001305 (rank : 162) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
TLR1_HUMAN
|
||||||
| NC score | 0.000951 (rank : 163) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15399, O15452, Q9UG90 | Gene names | TLR1, KIAA0012 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 1 precursor (Toll/interleukin-1 receptor-like protein) (TIL) (CD281 antigen). | |||||
|
AL2S2_MOUSE
|
||||||
| NC score | 0.000028 (rank : 164) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K4T3, Q6P922, Q8BU86, Q8BXK0, Q8K4T2 | Gene names | Als2cr2, Papk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ALS2CR2 (EC 2.7.11.1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 2 protein homolog) (ILP-interacting protein homolog) (Polyploidy-associated protein kinase). | |||||
|
PCDH9_HUMAN
|
||||||
| NC score | -0.000139 (rank : 165) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HC56 | Gene names | PCDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-9 precursor. | |||||
|
AL2S2_HUMAN
|
||||||
| NC score | -0.000626 (rank : 166) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9C0K7, Q5BKY7, Q9P1L0 | Gene names | ALS2CR2, ILPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ALS2CR2 (EC 2.7.11.1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 2 protein) (ILP- interacting protein) (CALS-21). | |||||
|
ZN286_HUMAN
|
||||||
| NC score | -0.002350 (rank : 167) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBT8, Q96JF3 | Gene names | ZNF286, KIAA1874 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 286. | |||||
|
ZN485_HUMAN
|
||||||
| NC score | -0.003048 (rank : 168) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NCK3, Q96CL0 | Gene names | ZNF485 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 485. | |||||