Please be patient as the page loads
|
CERU_MOUSE
|
||||||
| SwissProt Accessions | Q61147 | Gene names | Cp | |||
|
Domain Architecture |

|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CERU_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998657 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P00450, Q14063, Q2PP18, Q9UKS4 | Gene names | CP | |||
|
Domain Architecture |
|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
CERU_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61147 | Gene names | Cp | |||
|
Domain Architecture |

|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
HEPH_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.986895 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQS7, O75180, Q6UW45, Q9C058 | Gene names | HEPH, KIAA0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
HEPH_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.987103 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0Z4, Q6ZQ65, Q80Y80, Q8C4S2 | Gene names | Heph, Kiaa0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
FA8_HUMAN
|
||||||
| θ value | 1.02895e-112 (rank : 5) | NC score | 0.694526 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
FA8_MOUSE
|
||||||
| θ value | 4.32305e-111 (rank : 6) | NC score | 0.695125 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
FA5_HUMAN
|
||||||
| θ value | 6.45994e-107 (rank : 7) | NC score | 0.669355 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
CP2J6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 8) | NC score | 0.005832 (rank : 47) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54750 | Gene names | Cyp2j6 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2J6 (EC 1.14.14.1) (CYPIIJ6) (Arachidonic acid epoxygenase). | |||||
|
ZNT1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.031091 (rank : 32) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6M5, Q9BZF6 | Gene names | SLC30A1, ZNT1 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc transporter 1 (ZnT-1) (Solute carrier family 30 member 1). | |||||
|
CAN11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 10) | NC score | 0.014467 (rank : 37) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UMQ6, Q8N4R5 | Gene names | CAPN11 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-11 (EC 3.4.22.-) (Calcium-activated neutral proteinase 11) (CANP 11). | |||||
|
EGLN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.017643 (rank : 34) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
EGLN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.017411 (rank : 36) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
ZNT1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.027892 (rank : 33) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60738 | Gene names | Slc30a1, Znt1 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc transporter 1 (ZnT-1) (Solute carrier family 30 member 1). | |||||
|
ARY1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.011843 (rank : 40) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50294 | Gene names | Nat1, Aac1 | |||
|
Domain Architecture |

|
|||||
| Description | Arylamine N-acetyltransferase 1 (EC 2.3.1.5) (Arylamide acetylase 1) (N-acetyltransferase type 1) (NAT-1). | |||||
|
PHLD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.017447 (rank : 35) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P80108, Q15128 | Gene names | GPLD1, PIGPLD1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 1 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
IL1F5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.011863 (rank : 39) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QYY1, Q9JIG2 | Gene names | Il1f5, Fil1d, Il1h3, Il1hy1 | |||
|
Domain Architecture |
|
|||||
| Description | Interleukin-1 family member 5 (IL-1F5) (Interleukin-1 delta) (IL-1 delta) (Interleukin-1-like protein 1) (IL-1L1) (Interleukin-1 HY1) (IL-1HY1) (Interleukin-1 homolog 3) (IL-1H3). | |||||
|
ITPR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.008637 (rank : 41) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ITPR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.008633 (rank : 42) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
PHLD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.012741 (rank : 38) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15127 | Gene names | GPLD2, PIGPLD2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 2 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
CAN7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.008422 (rank : 43) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.008056 (rank : 44) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.007859 (rank : 46) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.007924 (rank : 45) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
CNTP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.063349 (rank : 30) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
CNTP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.061686 (rank : 31) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54991 | Gene names | Cntnap1, Nrxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (Paranodin) (NCP1) (MHDNIV). | |||||
|
CNTP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.071695 (rank : 25) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHC6, Q52LV1, Q5H9Q7, Q9UQ12 | Gene names | CNTNAP2, CASPR2, KIAA0868 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.067696 (rank : 29) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CPW0, Q6P2K4, Q6ZQ31 | Gene names | Cntnap2, Kiaa0868 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.068452 (rank : 28) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
CNTP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.069616 (rank : 27) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.070068 (rank : 26) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99P47 | Gene names | Cntnap4, Caspr4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CPXM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.077628 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96SM3, Q6P4G8, Q6UW65, Q9NUB5 | Gene names | CPXM1, CPX1, CPXM | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CPXM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.077156 (rank : 24) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z100, Q99LA3 | Gene names | Cpxm1, Cpx1, Cpxm | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CPXM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.091480 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N436 | Gene names | CPXM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CPXM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.092284 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D2L5, O54860, Q8VDQ4 | Gene names | Cpxm2, Cpx2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
DCBD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.128737 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DCBD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.116361 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
DCBD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.117942 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |
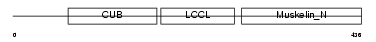
|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
EDIL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.135407 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
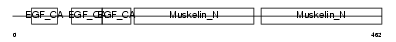
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EDIL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.132850 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
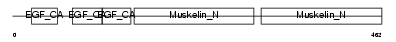
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
MFGM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.185042 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08431 | Gene names | MFGE8 | |||
|
Domain Architecture |
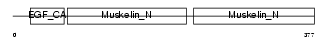
|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin]. | |||||
|
MFGM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.164292 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |
|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
NRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.105493 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
NRP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.107888 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
NRP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.108317 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.109308 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
XLRS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.152310 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15537 | Gene names | RS1, XLRS1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein). | |||||
|
XLRS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.152992 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1L4 | Gene names | Rs1, Rs1h, Xlrs1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein homolog). | |||||
|
CERU_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61147 | Gene names | Cp | |||
|
Domain Architecture |

|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
CERU_HUMAN
|
||||||
| NC score | 0.998657 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P00450, Q14063, Q2PP18, Q9UKS4 | Gene names | CP | |||
|
Domain Architecture |
|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
HEPH_MOUSE
|
||||||
| NC score | 0.987103 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0Z4, Q6ZQ65, Q80Y80, Q8C4S2 | Gene names | Heph, Kiaa0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
HEPH_HUMAN
|
||||||
| NC score | 0.986895 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQS7, O75180, Q6UW45, Q9C058 | Gene names | HEPH, KIAA0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
FA8_MOUSE
|
||||||
| NC score | 0.695125 (rank : 5) | θ value | 4.32305e-111 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
FA8_HUMAN
|
||||||
| NC score | 0.694526 (rank : 6) | θ value | 1.02895e-112 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
FA5_HUMAN
|
||||||
| NC score | 0.669355 (rank : 7) | θ value | 6.45994e-107 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
MFGM_HUMAN
|
||||||
| NC score | 0.185042 (rank : 8) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08431 | Gene names | MFGE8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin]. | |||||
|
MFGM_MOUSE
|
||||||
| NC score | 0.164292 (rank : 9) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
XLRS1_MOUSE
|
||||||
| NC score | 0.152992 (rank : 10) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1L4 | Gene names | Rs1, Rs1h, Xlrs1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein homolog). | |||||
|
XLRS1_HUMAN
|
||||||
| NC score | 0.152310 (rank : 11) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15537 | Gene names | RS1, XLRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein). | |||||
|
EDIL3_HUMAN
|
||||||
| NC score | 0.135407 (rank : 12) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
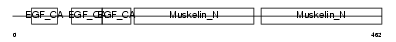
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EDIL3_MOUSE
|
||||||
| NC score | 0.132850 (rank : 13) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
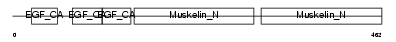
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
DCBD1_HUMAN
|
||||||
| NC score | 0.128737 (rank : 14) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DCBD2_MOUSE
|
||||||
| NC score | 0.117942 (rank : 15) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
DCBD2_HUMAN
|
||||||
| NC score | 0.116361 (rank : 16) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
NRP2_MOUSE
|
||||||
| NC score | 0.109308 (rank : 17) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP2_HUMAN
|
||||||
| NC score | 0.108317 (rank : 18) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP1_MOUSE
|
||||||
| NC score | 0.107888 (rank : 19) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
NRP1_HUMAN
|
||||||
| NC score | 0.105493 (rank : 20) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
CPXM2_MOUSE
|
||||||
| NC score | 0.092284 (rank : 21) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D2L5, O54860, Q8VDQ4 | Gene names | Cpxm2, Cpx2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CPXM2_HUMAN
|
||||||
| NC score | 0.091480 (rank : 22) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N436 | Gene names | CPXM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carboxypeptidase-like protein X2 precursor. | |||||
|
CPXM1_HUMAN
|
||||||
| NC score | 0.077628 (rank : 23) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96SM3, Q6P4G8, Q6UW65, Q9NUB5 | Gene names | CPXM1, CPX1, CPXM | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CPXM1_MOUSE
|
||||||
| NC score | 0.077156 (rank : 24) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z100, Q99LA3 | Gene names | Cpxm1, Cpx1, Cpxm | |||
|
Domain Architecture |

|
|||||
| Description | Probable carboxypeptidase X1 precursor (EC 3.4.17.-) (Metallocarboxypeptidase CPX-1). | |||||
|
CNTP2_HUMAN
|
||||||
| NC score | 0.071695 (rank : 25) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHC6, Q52LV1, Q5H9Q7, Q9UQ12 | Gene names | CNTNAP2, CASPR2, KIAA0868 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP4_MOUSE
|
||||||
| NC score | 0.070068 (rank : 26) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99P47 | Gene names | Cntnap4, Caspr4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP4_HUMAN
|
||||||
| NC score | 0.069616 (rank : 27) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CNTP3_HUMAN
|
||||||
| NC score | 0.068452 (rank : 28) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
CNTP2_MOUSE
|
||||||
| NC score | 0.067696 (rank : 29) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CPW0, Q6P2K4, Q6ZQ31 | Gene names | Cntnap2, Kiaa0868 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
CNTP1_HUMAN
|
||||||
| NC score | 0.063349 (rank : 30) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
CNTP1_MOUSE
|
||||||
| NC score | 0.061686 (rank : 31) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54991 | Gene names | Cntnap1, Nrxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (Paranodin) (NCP1) (MHDNIV). | |||||
|
ZNT1_HUMAN
|
||||||
| NC score | 0.031091 (rank : 32) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6M5, Q9BZF6 | Gene names | SLC30A1, ZNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc transporter 1 (ZnT-1) (Solute carrier family 30 member 1). | |||||
|
ZNT1_MOUSE
|
||||||
| NC score | 0.027892 (rank : 33) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60738 | Gene names | Slc30a1, Znt1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc transporter 1 (ZnT-1) (Solute carrier family 30 member 1). | |||||
|
EGLN1_HUMAN
|
||||||
| NC score | 0.017643 (rank : 34) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
PHLD1_HUMAN
|
||||||
| NC score | 0.017447 (rank : 35) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P80108, Q15128 | Gene names | GPLD1, PIGPLD1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 1 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
EGLN1_MOUSE
|
||||||
| NC score | 0.017411 (rank : 36) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
CAN11_HUMAN
|
||||||
| NC score | 0.014467 (rank : 37) | θ value | 1.81305 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UMQ6, Q8N4R5 | Gene names | CAPN11 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-11 (EC 3.4.22.-) (Calcium-activated neutral proteinase 11) (CANP 11). | |||||
|
PHLD2_HUMAN
|
||||||
| NC score | 0.012741 (rank : 38) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15127 | Gene names | GPLD2, PIGPLD2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 2 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
IL1F5_MOUSE
|
||||||
| NC score | 0.011863 (rank : 39) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QYY1, Q9JIG2 | Gene names | Il1f5, Fil1d, Il1h3, Il1hy1 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 family member 5 (IL-1F5) (Interleukin-1 delta) (IL-1 delta) (Interleukin-1-like protein 1) (IL-1L1) (Interleukin-1 HY1) (IL-1HY1) (Interleukin-1 homolog 3) (IL-1H3). | |||||
|
ARY1_MOUSE
|
||||||
| NC score | 0.011843 (rank : 40) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50294 | Gene names | Nat1, Aac1 | |||
|
Domain Architecture |

|
|||||
| Description | Arylamine N-acetyltransferase 1 (EC 2.3.1.5) (Arylamide acetylase 1) (N-acetyltransferase type 1) (NAT-1). | |||||
|
ITPR1_HUMAN
|
||||||
| NC score | 0.008637 (rank : 41) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ITPR1_MOUSE
|
||||||
| NC score | 0.008633 (rank : 42) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
CAN7_HUMAN
|
||||||
| NC score | 0.008422 (rank : 43) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.008056 (rank : 44) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.007924 (rank : 45) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.007859 (rank : 46) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
CP2J6_MOUSE
|
||||||
| NC score | 0.005832 (rank : 47) | θ value | 1.06291 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54750 | Gene names | Cyp2j6 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2J6 (EC 1.14.14.1) (CYPIIJ6) (Arachidonic acid epoxygenase). | |||||