Please be patient as the page loads
|
MAMC2_HUMAN
|
||||||
| SwissProt Accessions | Q7Z304, Q5VW47, Q8WX43, Q96BM4 | Gene names | MAMDC2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 2 precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MAMC2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z304, Q5VW47, Q8WX43, Q96BM4 | Gene names | MAMDC2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 2 precursor. | |||||
|
MAMC2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998292 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CG85, Q8BM24 | Gene names | Mamdc2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 2 precursor. | |||||
|
MAMC1_MOUSE
|
||||||
| θ value | 7.56453e-23 (rank : 3) | NC score | 0.372404 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P60755 | Gene names | Mamdc1, Mdga2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 1 precursor (MAM domain-containing glycosylphosphatidylinositol anchor protein 2). | |||||
|
MAMC1_HUMAN
|
||||||
| θ value | 9.87957e-23 (rank : 4) | NC score | 0.369282 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z553 | Gene names | MAMDC1, MDGA2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 1 precursor (MAM domain-containing glycosylphosphatidylinositol anchor protein 2). | |||||
|
MDGA1_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 5) | NC score | 0.376667 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NFP4, Q8NBE3 | Gene names | MDGA1, MAMDC3 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing glycosylphosphatidylinositol anchor protein 1 precursor (MAM domain-containing protein 3) (GPI and MAM protein) (Glycosylphosphatidylinositol-MAM) (GPIM). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 6) | NC score | 0.319468 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 7) | NC score | 0.242016 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ENTK_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 8) | NC score | 0.162821 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
NRP1_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 9) | NC score | 0.330437 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
NRP1_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 10) | NC score | 0.331430 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
ENTK_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 11) | NC score | 0.160622 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
PTPRU_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 12) | NC score | 0.249531 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92729 | Gene names | PTPRU, PCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase U precursor (EC 3.1.3.48) (R-PTP-U) (Protein-tyrosine phosphatase J) (PTP-J) (Pancreatic carcinoma phosphatase 2) (PCP-2). | |||||
|
PTPRT_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 13) | NC score | 0.239567 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14522, O43655, O75664, Q5W0X9, Q5W0Y1, Q9BR24, Q9BR28, Q9H0Y8, Q9NTL1, Q9NU72, Q9UBD2, Q9UJL7 | Gene names | PTPRT, KIAA0283 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho). | |||||
|
PTPRM_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 14) | NC score | 0.242924 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P28827 | Gene names | PTPRM, PTPRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRK_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 15) | NC score | 0.263547 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRM_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 16) | NC score | 0.235763 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P28828 | Gene names | Ptprm | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRK_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 17) | NC score | 0.261928 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
MEP1B_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 18) | NC score | 0.337127 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61847 | Gene names | Mep1b, Mep-1b | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit beta precursor (EC 3.4.24.18) (Endopeptidase-2). | |||||
|
PTPRT_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 19) | NC score | 0.235459 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
MEP1A_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 20) | NC score | 0.338456 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P28825 | Gene names | Mep1a | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit alpha precursor (EC 3.4.24.18) (Endopeptidase-2) (MEP-1). | |||||
|
NRP2_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 21) | NC score | 0.260064 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP2_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 22) | NC score | 0.248238 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
MEP1B_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 23) | NC score | 0.343849 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16820, Q670J1 | Gene names | MEP1B | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit beta precursor (EC 3.4.24.18) (Endopeptidase-2) (N- benzoyl-L-tyrosyl-P-amino-benzoic acid hydrolase subunit beta) (PABA peptide hydrolase) (PPH beta). | |||||
|
MEP1A_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 24) | NC score | 0.314254 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16819 | Gene names | MEP1A | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit alpha precursor (EC 3.4.24.18) (Endopeptidase-2) (N- benzoyl-L-tyrosyl-P-amino-benzoic acid hydrolase subunit alpha) (PABA peptide hydrolase) (PPH alpha). | |||||
|
PC11Y_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.008620 (rank : 166) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZA8, Q70LR6, Q70LR8, Q70LS0, Q70LS1, Q70LS2, Q70LS3, Q70LS4, Q70LS5, Q8WY34, Q9BZA9, Q9H4E1 | Gene names | PCDH11Y, PCDH11, PCDH22, PCDHY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 Y-linked precursor (Protocadherin-11) (Protocadherin- 22) (Protocadherin on the Y chromosome) (PCDH-Y) (Protocadherin prostate cancer) (Protocadherin-PC). | |||||
|
TAF1C_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.019655 (rank : 162) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PDZ2, P97359, Q3U3B6, Q8BN54 | Gene names | Taf1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor, RNA polymerase I, subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 95 kDa) (TAFI95). | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.012382 (rank : 163) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
ADA19_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.005045 (rank : 167) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H013, Q9BZL5, Q9UHP2 | Gene names | ADAM19, MLTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta) (Metalloprotease and disintegrin dentritic antigen marker) (MADDAM). | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.011738 (rank : 164) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
ZN546_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | -0.001763 (rank : 168) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UE3 | Gene names | ZNF546, ZNF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 546. | |||||
|
PRKDC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.010528 (rank : 165) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
ASTL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.078671 (rank : 90) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
ASTL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.089846 (rank : 72) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6HA09, Q8BMA1 | Gene names | Astl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
BMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.092762 (rank : 59) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
BMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.093174 (rank : 56) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
BMPER_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.064265 (rank : 120) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
BMPER_MOUSE
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.063113 (rank : 122) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
CD45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.111462 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
CD45_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.108339 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
CNTN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.057700 (rank : 139) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12860, Q12861, Q14030, Q8N466 | Gene names | CNTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3) (Glycoprotein gp135). | |||||
|
CNTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.059016 (rank : 133) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P12960, Q6NXV7, Q8BR42, Q8C6A0 | Gene names | Cntn1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3). | |||||
|
CNTN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.062734 (rank : 124) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02246, P78432 | Gene names | CNTN2, TAG1, TAX1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-2 precursor (Axonin-1) (Axonal glycoprotein TAG-1) (Transient axonal glycoprotein 1) (TAX-1). | |||||
|
CNTN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.063035 (rank : 123) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61330, Q6NZJ4, Q7TSJ5 | Gene names | Cntn2, Tax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-2 precursor (Axonin-1) (Axonal glycoprotein TAG-1) (Transient axonal glycoprotein 1) (TAX-1). | |||||
|
CNTN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.058148 (rank : 136) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P232, Q9H039 | Gene names | CNTN3, KIAA1496, PANG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-3 precursor (Brain-derived immunoglobulin superfamily protein 1) (BIG-1) (Plasmacytoma-associated neuronal glycoprotein). | |||||
|
CNTN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.061419 (rank : 127) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07409 | Gene names | Cntn3, Pang, Pcs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-3 precursor (Brain-derived immunoglobulin superfamily protein 1) (BIG-1) (Plasmacytoma-associated neuronal glycoprotein). | |||||
|
CNTN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.059214 (rank : 132) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IWV2, Q8IX14, Q8TC35 | Gene names | CNTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-4 precursor (Brain-derived immunoglobulin superfamily protein 2) (BIG-2). | |||||
|
CNTN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.056381 (rank : 142) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q69Z26, Q8BRT6, Q8CBD3 | Gene names | Cntn4, Kiaa3024 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-4 precursor (Brain-derived immunoglobulin superfamily protein 2) (BIG-2). | |||||
|
CNTN5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.068858 (rank : 112) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94779, O94780 | Gene names | CNTN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-5 precursor (Neural recognition molecule NB-2) (hNB-2). | |||||
|
CNTN5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.069426 (rank : 110) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P68500 | Gene names | Cntn5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-5 precursor (Neural recognition molecule NB-2). | |||||
|
CNTN6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.056592 (rank : 141) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQ52 | Gene names | CNTN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (hNB-3). | |||||
|
CNTN6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.052764 (rank : 150) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JMB8, Q8C6X1 | Gene names | Cntn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (mNB-3). | |||||
|
CSMD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.051244 (rank : 158) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CSMD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.050879 (rank : 160) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.052408 (rank : 151) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
CUBN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.054752 (rank : 144) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
CUBN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.054074 (rank : 146) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
CUZD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.076075 (rank : 94) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUZD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.075704 (rank : 95) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
DCBD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.074416 (rank : 97) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DCBD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.078091 (rank : 91) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
DCBD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.081347 (rank : 83) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |
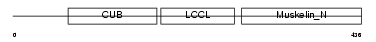
|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
DCC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.061790 (rank : 126) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
DCC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.062368 (rank : 125) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
DMBT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.058014 (rank : 137) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
DSCAM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.058770 (rank : 134) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60469, O60468 | Gene names | DSCAM | |||
|
Domain Architecture |

|
|||||
| Description | Down syndrome cell adhesion molecule precursor (CHD2). | |||||
|
DSCL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.059577 (rank : 131) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
EDIL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.069697 (rank : 107) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
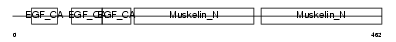
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EDIL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.069621 (rank : 108) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
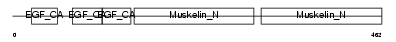
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
FA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.064774 (rank : 119) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
FA8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.068599 (rank : 115) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
FA8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.068641 (rank : 114) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
L1CAM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.057796 (rank : 138) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P32004, Q8TA87 | Gene names | L1CAM, CAML1, MIC5 | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule L1 precursor (N-CAM L1) (CD171 antigen). | |||||
|
L1CAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.054361 (rank : 145) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P11627 | Gene names | L1cam, Caml1 | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule L1 precursor (N-CAM L1) (CD171 antigen). | |||||
|
LCE3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.085574 (rank : 79) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TA76 | Gene names | LCE3A, LEP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3A (Late envelope protein 13). | |||||
|
LRP12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.050618 (rank : 161) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |
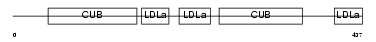
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
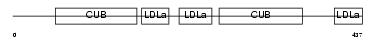
LRP12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.052210 (rank : 153) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
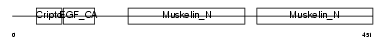
MFGM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.097025 (rank : 40) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08431 | Gene names | MFGE8 | |||
|
Domain Architecture |
|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin]. | |||||
|
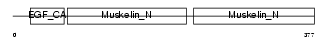
MFGM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.078948 (rank : 88) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |
|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
MFRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.068990 (rank : 111) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MFRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.068796 (rank : 113) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.052279 (rank : 152) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.056182 (rank : 143) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
NEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.058376 (rank : 135) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92859, O00340 | Gene names | NEO1, NGN | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
NEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.059588 (rank : 130) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97798 | Gene names | Neo1, Ngn | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
NETO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.077053 (rank : 93) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.079518 (rank : 87) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.081712 (rank : 82) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NETO2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.079576 (rank : 86) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NFASC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.057599 (rank : 140) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94856, Q5T2F0, Q5T2F1, Q5T2F2, Q5T2F3, Q5T2F4, Q5T2F5, Q5T2F6, Q5T2F7, Q5T2F9, Q5T2G0, Q5W9F8, Q68DH3, Q6ZQV6, Q7Z3K1, Q96HT1, Q96K50 | Gene names | NFASC, KIAA0756 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
NFASC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.059965 (rank : 128) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q810U3 | Gene names | Nfasc | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
NRCAM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.063347 (rank : 121) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92823, O15051, O15179, Q9UHI3, Q9UHI4 | Gene names | NRCAM, KIAA0343 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal cell adhesion molecule precursor (Nr-CAM) (NgCAM-related cell adhesion molecule) (Ng-CAM-related) (hBravo). | |||||
|
NRCAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.059894 (rank : 129) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q810U4, Q80U33, Q8BLG8, Q8BX92, Q8BYJ8 | Gene names | Nrcam, Kiaa0343 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal cell adhesion molecule precursor (Nr-CAM) (NgCAM-related cell adhesion molecule) (Ng-CAM-related) (mBravo). | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.082166 (rank : 81) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
PCOC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.082805 (rank : 80) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
PCOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.088402 (rank : 74) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PCOC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.090023 (rank : 71) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PDGFD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.075478 (rank : 96) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9GZP0, Q9BWV5 | Gene names | PDGFD, IEGF, SCDGFB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor D precursor (PDGF D) (Iris-expressed growth factor) (Spinal cord-derived growth factor B) (SCDGF-B). | |||||
|
PDGFD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.068305 (rank : 116) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q925I7, Q3URF6, Q8K2L3, Q9D1L8 | Gene names | Pdgfd, Scdgfb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor chain precursor (PDGF D) (Spinal cord- derived growth factor B) (SCDGF-B). | |||||
|
PTN11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.087259 (rank : 76) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06124, Q96HD7 | Gene names | PTPN11, PTP2C, SHPTP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase 2C) (PTP-2C) (PTP-1D) (SH-PTP3) (SH- PTP2) (SHP-2) (Shp2). | |||||
|
PTN11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.087270 (rank : 75) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35235, Q3TQ84, Q64509, Q6PCL5 | Gene names | Ptpn11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase SYP) (SH-PTP2) (SHP-2) (Shp2). | |||||
|
PTN12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.093795 (rank : 49) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
PTN12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.094070 (rank : 47) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.066841 (rank : 118) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.067004 (rank : 117) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
PTN14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.070396 (rank : 104) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
PTN14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.069848 (rank : 106) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
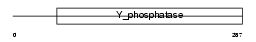
PTN18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.089018 (rank : 73) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99952 | Gene names | PTPN18, BDP1 | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Brain-derived phosphatase). | |||||
|
PTN18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.091797 (rank : 63) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
PTN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.091054 (rank : 67) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18031, Q5TGD8, Q9BQV9, Q9NQQ4 | Gene names | PTPN1, PTP1B | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B). | |||||
|
PTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.091002 (rank : 68) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35821, Q60840, Q62131, Q64498, Q99JS1 | Gene names | Ptpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B) (Protein-protein-tyrosine phosphatase HA2) (PTP-HA2). | |||||
|
PTN21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.069869 (rank : 105) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
PTN21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.069437 (rank : 109) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
PTN22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.092450 (rank : 61) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
PTN22_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.091547 (rank : 66) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.071096 (rank : 102) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.071796 (rank : 99) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PTN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.093106 (rank : 57) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17706, Q96HR2 | Gene names | PTPN2, PTPT | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (T-cell protein-tyrosine phosphatase) (TCPTP). | |||||
|
PTN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.091592 (rank : 65) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06180, Q3V259, Q922E7 | Gene names | Ptpn2, Ptpt | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-2) (MPTP). | |||||
|
PTN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.071204 (rank : 101) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
PTN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.070665 (rank : 103) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
PTN5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.096529 (rank : 41) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54829, Q8N2A1 | Gene names | PTPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.097246 (rank : 39) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.087217 (rank : 77) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29350, Q969V8 | Gene names | PTPN6, HCP, PTP1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (SH-PTP1) (Protein-tyrosine phosphatase SHP-1). | |||||
|
PTN6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.087003 (rank : 78) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29351, O35128, Q63872, Q63873, Q63874, Q921G3, Q9QVA6, Q9QVA7, Q9QVA8, Q9R0V6 | Gene names | Ptpn6, Hcp, Hcph, Ptp1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (70Z-SHP) (SH-PTP1) (SHP-1) (PTPTY-42). | |||||
|
PTN7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.094933 (rank : 45) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35236, Q9BV05 | Gene names | PTPN7 | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48) (Protein-tyrosine phosphatase LC-PTP) (Hematopoietic protein-tyrosine phosphatase) (HEPTP). | |||||
|
PTN7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.095453 (rank : 43) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BUM3 | Gene names | Ptpn7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48). | |||||
|
PTN9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.092562 (rank : 60) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43378 | Gene names | PTPN9 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.093293 (rank : 52) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35239, Q7TSK0 | Gene names | Ptpn9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTPR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.092839 (rank : 58) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92932, Q8N4I5, Q92662 | Gene names | PTPRN2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (Islet cell autoantigen-related protein) (ICAAR) (IAR) (Phogrin). | |||||
|
PTPR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.092052 (rank : 62) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80560, O09134, P70328 | Gene names | Ptprn2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (PTP IA-2beta) (Protein tyrosine phosphatase-NP) (PTP-NP). | |||||
|
PTPRA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.115418 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
PTPRA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.114133 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18052, Q61808 | Gene names | Ptpra, Lrp, Ptpa | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha) (LCA- related phosphatase) (PTPTY-28). | |||||
|
PTPRB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.091628 (rank : 64) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23467 | Gene names | PTPRB, PTPB | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase beta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase beta) (R-PTP-beta). | |||||
|
PTPRD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.098406 (rank : 37) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.098349 (rank : 38) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.115510 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23469, Q96KQ6 | Gene names | PTPRE | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.115798 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49446, Q62134, Q62444, Q64496 | Gene names | Ptpre, Ptpe | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.099912 (rank : 33) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10586 | Gene names | PTPRF, LAR | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase F precursor (EC 3.1.3.48) (LAR protein) (Leukocyte antigen related). | |||||
|
PTPRG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.099098 (rank : 35) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.098728 (rank : 36) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRJ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.093987 (rank : 48) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
PTPRJ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.094540 (rank : 46) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PTPRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.090405 (rank : 69) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.090110 (rank : 70) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
PTPRO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.099877 (rank : 34) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
PTPRR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.093732 (rank : 50) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
PTPRR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.093181 (rank : 55) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62132, Q64491, Q64492, Q9QUH9 | Gene names | Ptprr, Ptp13 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine-phosphatase SL) (Phosphotyrosine phosphatase 13). | |||||
|
PTPRS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.100680 (rank : 32) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13332, O75255, O75870, Q15718, Q16341 | Gene names | PTPRS | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase S precursor (EC 3.1.3.48) (R-PTP-S) (Protein-tyrosine phosphatase sigma) (R-PTP-sigma). | |||||
|
PTPRV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.093287 (rank : 53) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.101670 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
SDK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.051485 (rank : 157) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z5N4, Q8TEN9, Q8TEP5, Q96N44 | Gene names | SDK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-1 precursor. | |||||
|
SDK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.053299 (rank : 148) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3UH53, Q3UFD1, Q6PAL2, Q6V3A4, Q8BMC2, Q8BZI1 | Gene names | Sdk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-1 precursor. | |||||
|
SDK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.050885 (rank : 159) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q58EX2, Q86VY3, Q9NTD2, Q9NVB3, Q9P214 | Gene names | SDK2, KIAA1514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-2 precursor. | |||||
|
SDK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.051993 (rank : 154) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6V4S5, Q3TTK1, Q5U5W7, Q6ZPP2 | Gene names | Sdk2, Kiaa1514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-2 precursor. | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.051534 (rank : 156) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
SSPO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.051956 (rank : 155) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
TECTA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.080801 (rank : 84) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75443 | Gene names | TECTA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TECTA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.080272 (rank : 85) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08523 | Gene names | Tecta | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.093650 (rank : 51) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
TLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.093285 (rank : 54) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
TLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.096341 (rank : 42) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TLL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.095049 (rank : 44) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TSG6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.077787 (rank : 92) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TSG6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.078880 (rank : 89) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
VWF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.052833 (rank : 149) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
VWF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.053935 (rank : 147) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
XLRS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.073625 (rank : 98) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15537 | Gene names | RS1, XLRS1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein). | |||||
|
XLRS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.071325 (rank : 100) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1L4 | Gene names | Rs1, Rs1h, Xlrs1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein homolog). | |||||
|
MAMC2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z304, Q5VW47, Q8WX43, Q96BM4 | Gene names | MAMDC2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 2 precursor. | |||||
|
MAMC2_MOUSE
|
||||||
| NC score | 0.998292 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CG85, Q8BM24 | Gene names | Mamdc2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 2 precursor. | |||||
|
MDGA1_HUMAN
|
||||||
| NC score | 0.376667 (rank : 3) | θ value | 8.65492e-19 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NFP4, Q8NBE3 | Gene names | MDGA1, MAMDC3 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing glycosylphosphatidylinositol anchor protein 1 precursor (MAM domain-containing protein 3) (GPI and MAM protein) (Glycosylphosphatidylinositol-MAM) (GPIM). | |||||
|
MAMC1_MOUSE
|
||||||
| NC score | 0.372404 (rank : 4) | θ value | 7.56453e-23 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P60755 | Gene names | Mamdc1, Mdga2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 1 precursor (MAM domain-containing glycosylphosphatidylinositol anchor protein 2). | |||||
|
MAMC1_HUMAN
|
||||||
| NC score | 0.369282 (rank : 5) | θ value | 9.87957e-23 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z553 | Gene names | MAMDC1, MDGA2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 1 precursor (MAM domain-containing glycosylphosphatidylinositol anchor protein 2). | |||||
|
MEP1B_HUMAN
|
||||||
| NC score | 0.343849 (rank : 6) | θ value | 4.92598e-06 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16820, Q670J1 | Gene names | MEP1B | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit beta precursor (EC 3.4.24.18) (Endopeptidase-2) (N- benzoyl-L-tyrosyl-P-amino-benzoic acid hydrolase subunit beta) (PABA peptide hydrolase) (PPH beta). | |||||
|
MEP1A_MOUSE
|
||||||
| NC score | 0.338456 (rank : 7) | θ value | 2.61198e-07 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P28825 | Gene names | Mep1a | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit alpha precursor (EC 3.4.24.18) (Endopeptidase-2) (MEP-1). | |||||
|
MEP1B_MOUSE
|
||||||
| NC score | 0.337127 (rank : 8) | θ value | 3.08544e-08 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61847 | Gene names | Mep1b, Mep-1b | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit beta precursor (EC 3.4.24.18) (Endopeptidase-2). | |||||
|
NRP1_HUMAN
|
||||||
| NC score | 0.331430 (rank : 9) | θ value | 4.16044e-13 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
NRP1_MOUSE
|
||||||
| NC score | 0.330437 (rank : 10) | θ value | 3.18553e-13 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.319468 (rank : 11) | θ value | 1.92812e-18 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
MEP1A_HUMAN
|
||||||
| NC score | 0.314254 (rank : 12) | θ value | 5.44631e-05 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16819 | Gene names | MEP1A | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit alpha precursor (EC 3.4.24.18) (Endopeptidase-2) (N- benzoyl-L-tyrosyl-P-amino-benzoic acid hydrolase subunit alpha) (PABA peptide hydrolase) (PPH alpha). | |||||
|
PTPRK_MOUSE
|
||||||
| NC score | 0.263547 (rank : 13) | θ value | 1.06045e-08 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRK_HUMAN
|
||||||
| NC score | 0.261928 (rank : 14) | θ value | 1.80886e-08 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
NRP2_HUMAN
|
||||||
| NC score | 0.260064 (rank : 15) | θ value | 2.61198e-07 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
PTPRU_HUMAN
|
||||||
| NC score | 0.249531 (rank : 16) | θ value | 2.79066e-09 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92729 | Gene names | PTPRU, PCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase U precursor (EC 3.1.3.48) (R-PTP-U) (Protein-tyrosine phosphatase J) (PTP-J) (Pancreatic carcinoma phosphatase 2) (PCP-2). | |||||
|
NRP2_MOUSE
|
||||||
| NC score | 0.248238 (rank : 17) | θ value | 5.81887e-07 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
PTPRM_HUMAN
|
||||||
| NC score | 0.242924 (rank : 18) | θ value | 8.11959e-09 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P28827 | Gene names | PTPRM, PTPRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.242016 (rank : 19) | θ value | 5.8054e-15 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
PTPRT_HUMAN
|
||||||
| NC score | 0.239567 (rank : 20) | θ value | 6.21693e-09 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14522, O43655, O75664, Q5W0X9, Q5W0Y1, Q9BR24, Q9BR28, Q9H0Y8, Q9NTL1, Q9NU72, Q9UBD2, Q9UJL7 | Gene names | PTPRT, KIAA0283 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho). | |||||
|
PTPRM_MOUSE
|
||||||
| NC score | 0.235763 (rank : 21) | θ value | 1.06045e-08 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P28828 | Gene names | Ptprm | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRT_MOUSE
|
||||||
| NC score | 0.235459 (rank : 22) | θ value | 1.17247e-07 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
ENTK_HUMAN
|
||||||
| NC score | 0.162821 (rank : 23) | θ value | 3.76295e-14 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
ENTK_MOUSE
|
||||||
| NC score | 0.160622 (rank : 24) | θ value | 4.59992e-12 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
PTPRE_MOUSE
|
||||||
| NC score | 0.115798 (rank : 25) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49446, Q62134, Q62444, Q64496 | Gene names | Ptpre, Ptpe | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRE_HUMAN
|
||||||
| NC score | 0.115510 (rank : 26) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23469, Q96KQ6 | Gene names | PTPRE | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRA_HUMAN
|
||||||
| NC score | 0.115418 (rank : 27) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
PTPRA_MOUSE
|
||||||
| NC score | 0.114133 (rank : 28) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18052, Q61808 | Gene names | Ptpra, Lrp, Ptpa | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha) (LCA- related phosphatase) (PTPTY-28). | |||||
|
CD45_HUMAN
|
||||||
| NC score | 0.111462 (rank : 29) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
CD45_MOUSE
|
||||||
| NC score | 0.108339 (rank : 30) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.101670 (rank : 31) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
PTPRS_HUMAN
|
||||||
| NC score | 0.100680 (rank : 32) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13332, O75255, O75870, Q15718, Q16341 | Gene names | PTPRS | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase S precursor (EC 3.1.3.48) (R-PTP-S) (Protein-tyrosine phosphatase sigma) (R-PTP-sigma). | |||||
|
PTPRF_HUMAN
|
||||||
| NC score | 0.099912 (rank : 33) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10586 | Gene names | PTPRF, LAR | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase F precursor (EC 3.1.3.48) (LAR protein) (Leukocyte antigen related). | |||||
|
PTPRO_HUMAN
|
||||||
| NC score | 0.099877 (rank : 34) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
PTPRG_HUMAN
|
||||||
| NC score | 0.099098 (rank : 35) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRG_MOUSE
|
||||||
| NC score | 0.098728 (rank : 36) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRD_HUMAN
|
||||||
| NC score | 0.098406 (rank : 37) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRD_MOUSE
|
||||||
| NC score | 0.098349 (rank : 38) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTN5_MOUSE
|
||||||
| NC score | 0.097246 (rank : 39) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
MFGM_HUMAN
|
||||||
| NC score | 0.097025 (rank : 40) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08431 | Gene names | MFGE8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (HMFG) (Breast epithelial antigen BA46) (MFGM) [Contains: Lactadherin short form; Medin]. | |||||
|
PTN5_HUMAN
|
||||||
| NC score | 0.096529 (rank : 41) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54829, Q8N2A1 | Gene names | PTPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
TLL2_HUMAN
|
||||||
| NC score | 0.096341 (rank : 42) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
PTN7_MOUSE
|
||||||
| NC score | 0.095453 (rank : 43) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BUM3 | Gene names | Ptpn7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48). | |||||
|
TLL2_MOUSE
|
||||||
| NC score | 0.095049 (rank : 44) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
PTN7_HUMAN
|
||||||
| NC score | 0.094933 (rank : 45) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35236, Q9BV05 | Gene names | PTPN7 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48) (Protein-tyrosine phosphatase LC-PTP) (Hematopoietic protein-tyrosine phosphatase) (HEPTP). | |||||
|
PTPRJ_MOUSE
|
||||||
| NC score | 0.094540 (rank : 46) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PTN12_MOUSE
|
||||||
| NC score | 0.094070 (rank : 47) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
PTPRJ_HUMAN
|
||||||
| NC score | 0.093987 (rank : 48) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
PTN12_HUMAN
|
||||||
| NC score | 0.093795 (rank : 49) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
PTPRR_HUMAN
|
||||||
| NC score | 0.093732 (rank : 50) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
TLL1_HUMAN
|
||||||
| NC score | 0.093650 (rank : 51) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
PTN9_MOUSE
|
||||||
| NC score | 0.093293 (rank : 52) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35239, Q7TSK0 | Gene names | Ptpn9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTPRV_MOUSE
|
||||||
| NC score | 0.093287 (rank : 53) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
TLL1_MOUSE
|
||||||
| NC score | 0.093285 (rank : 54) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
PTPRR_MOUSE
|
||||||
| NC score | 0.093181 (rank : 55) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62132, Q64491, Q64492, Q9QUH9 | Gene names | Ptprr, Ptp13 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine-phosphatase SL) (Phosphotyrosine phosphatase 13). | |||||
|
BMP1_MOUSE
|
||||||
| NC score | 0.093174 (rank : 56) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
PTN2_HUMAN
|
||||||
| NC score | 0.093106 (rank : 57) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17706, Q96HR2 | Gene names | PTPN2, PTPT | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (T-cell protein-tyrosine phosphatase) (TCPTP). | |||||
|
PTPR2_HUMAN
|
||||||
| NC score | 0.092839 (rank : 58) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92932, Q8N4I5, Q92662 | Gene names | PTPRN2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (Islet cell autoantigen-related protein) (ICAAR) (IAR) (Phogrin). | |||||
|
BMP1_HUMAN
|
||||||
| NC score | 0.092762 (rank : 59) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
PTN9_HUMAN
|
||||||
| NC score | 0.092562 (rank : 60) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43378 | Gene names | PTPN9 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN22_HUMAN
|
||||||
| NC score | 0.092450 (rank : 61) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
PTPR2_MOUSE
|
||||||
| NC score | 0.092052 (rank : 62) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80560, O09134, P70328 | Gene names | Ptprn2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (PTP IA-2beta) (Protein tyrosine phosphatase-NP) (PTP-NP). | |||||
|
PTN18_MOUSE
|
||||||
| NC score | 0.091797 (rank : 63) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
PTPRB_HUMAN
|
||||||
| NC score | 0.091628 (rank : 64) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23467 | Gene names | PTPRB, PTPB | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase beta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase beta) (R-PTP-beta). | |||||
|
PTN2_MOUSE
|
||||||
| NC score | 0.091592 (rank : 65) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06180, Q3V259, Q922E7 | Gene names | Ptpn2, Ptpt | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-2) (MPTP). | |||||
|
PTN22_MOUSE
|
||||||
| NC score | 0.091547 (rank : 66) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
PTN1_HUMAN
|
||||||
| NC score | 0.091054 (rank : 67) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18031, Q5TGD8, Q9BQV9, Q9NQQ4 | Gene names | PTPN1, PTP1B | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B). | |||||
|
PTN1_MOUSE
|
||||||
| NC score | 0.091002 (rank : 68) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35821, Q60840, Q62131, Q64498, Q99JS1 | Gene names | Ptpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B) (Protein-protein-tyrosine phosphatase HA2) (PTP-HA2). | |||||
|
PTPRN_HUMAN
|
||||||
| NC score | 0.090405 (rank : 69) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.090110 (rank : 70) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
PCOC2_MOUSE
|
||||||
| NC score | 0.090023 (rank : 71) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
ASTL_MOUSE
|
||||||
| NC score | 0.089846 (rank : 72) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6HA09, Q8BMA1 | Gene names | Astl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
PTN18_HUMAN
|
||||||
| NC score | 0.089018 (rank : 73) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99952 | Gene names | PTPN18, BDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Brain-derived phosphatase). | |||||
|
PCOC2_HUMAN
|
||||||
| NC score | 0.088402 (rank : 74) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PTN11_MOUSE
|
||||||
| NC score | 0.087270 (rank : 75) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35235, Q3TQ84, Q64509, Q6PCL5 | Gene names | Ptpn11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase SYP) (SH-PTP2) (SHP-2) (Shp2). | |||||
|
PTN11_HUMAN
|
||||||
| NC score | 0.087259 (rank : 76) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06124, Q96HD7 | Gene names | PTPN11, PTP2C, SHPTP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase 2C) (PTP-2C) (PTP-1D) (SH-PTP3) (SH- PTP2) (SHP-2) (Shp2). | |||||
|
PTN6_HUMAN
|
||||||
| NC score | 0.087217 (rank : 77) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29350, Q969V8 | Gene names | PTPN6, HCP, PTP1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (SH-PTP1) (Protein-tyrosine phosphatase SHP-1). | |||||
|
PTN6_MOUSE
|
||||||
| NC score | 0.087003 (rank : 78) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29351, O35128, Q63872, Q63873, Q63874, Q921G3, Q9QVA6, Q9QVA7, Q9QVA8, Q9R0V6 | Gene names | Ptpn6, Hcp, Hcph, Ptp1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (70Z-SHP) (SH-PTP1) (SHP-1) (PTPTY-42). | |||||
|
LCE3A_HUMAN
|
||||||
| NC score | 0.085574 (rank : 79) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TA76 | Gene names | LCE3A, LEP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3A (Late envelope protein 13). | |||||
|
PCOC1_MOUSE
|
||||||
| NC score | 0.082805 (rank : 80) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.082166 (rank : 81) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
NETO2_HUMAN
|
||||||
| NC score | 0.081712 (rank : 82) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
DCBD2_MOUSE
|
||||||
| NC score | 0.081347 (rank : 83) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
TECTA_HUMAN
|
||||||
| NC score | 0.080801 (rank : 84) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75443 | Gene names | TECTA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TECTA_MOUSE
|
||||||
| NC score | 0.080272 (rank : 85) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08523 | Gene names | Tecta | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
NETO2_MOUSE
|
||||||
| NC score | 0.079576 (rank : 86) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NETO1_MOUSE
|
||||||
| NC score | 0.079518 (rank : 87) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
MFGM_MOUSE
|
||||||
| NC score | 0.078948 (rank : 88) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P21956, P97800, Q3TBN5, Q9R1X9, Q9WTS3 | Gene names | Mfge8 | |||
|
Domain Architecture |

|
|||||
| Description | Lactadherin precursor (Milk fat globule-EGF factor 8) (MFG-E8) (MFGM) (Sperm surface protein SP47) (MP47). | |||||
|
TSG6_MOUSE
|
||||||
| NC score | 0.078880 (rank : 89) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
ASTL_HUMAN
|
||||||
| NC score | 0.078671 (rank : 90) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
DCBD2_HUMAN
|
||||||
| NC score | 0.078091 (rank : 91) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
TSG6_HUMAN
|
||||||
| NC score | 0.077787 (rank : 92) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
NETO1_HUMAN
|
||||||
| NC score | 0.077053 (rank : 93) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
CUZD1_HUMAN
|
||||||
| NC score | 0.076075 (rank : 94) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUZD1_MOUSE
|
||||||
| NC score | 0.075704 (rank : 95) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
PDGFD_HUMAN
|
||||||
| NC score | 0.075478 (rank : 96) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9GZP0, Q9BWV5 | Gene names | PDGFD, IEGF, SCDGFB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor D precursor (PDGF D) (Iris-expressed growth factor) (Spinal cord-derived growth factor B) (SCDGF-B). | |||||
|
DCBD1_HUMAN
|
||||||
| NC score | 0.074416 (rank : 97) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
XLRS1_HUMAN
|
||||||
| NC score | 0.073625 (rank : 98) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15537 | Gene names | RS1, XLRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.071796 (rank : 99) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
XLRS1_MOUSE
|
||||||
| NC score | 0.071325 (rank : 100) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1L4 | Gene names | Rs1, Rs1h, Xlrs1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoschisin precursor (X-linked juvenile retinoschisis protein homolog). | |||||
|
PTN3_HUMAN
|
||||||
| NC score | 0.071204 (rank : 101) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.071096 (rank : 102) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
PTN4_HUMAN
|
||||||
| NC score | 0.070665 (rank : 103) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
PTN14_HUMAN
|
||||||
| NC score | 0.070396 (rank : 104) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
PTN21_HUMAN
|
||||||
| NC score | 0.069869 (rank : 105) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
PTN14_MOUSE
|
||||||
| NC score | 0.069848 (rank : 106) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
EDIL3_HUMAN
|
||||||
| NC score | 0.069697 (rank : 107) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43854, O43855 | Gene names | EDIL3, DEL1 | |||
|
Domain Architecture |
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
EDIL3_MOUSE
|
||||||
| NC score | 0.069621 (rank : 108) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35474, O35475 | Gene names | Edil3, Del1 | |||
|
Domain Architecture |
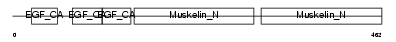
|
|||||
| Description | EGF-like repeat and discoidin I-like domain-containing protein 3 precursor (EGF-like repeats and discoidin I-like domains protein 3) (Developmentally-regulated endothelial cell locus 1 protein) (Integrin-binding protein DEL1). | |||||
|
PTN21_MOUSE
|
||||||
| NC score | 0.069437 (rank : 109) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
CNTN5_MOUSE
|
||||||
| NC score | 0.069426 (rank : 110) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P68500 | Gene names | Cntn5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-5 precursor (Neural recognition molecule NB-2). | |||||
|
MFRP_HUMAN
|
||||||
| NC score | 0.068990 (rank : 111) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
CNTN5_HUMAN
|
||||||
| NC score | 0.068858 (rank : 112) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94779, O94780 | Gene names | CNTN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-5 precursor (Neural recognition molecule NB-2) (hNB-2). | |||||
|
MFRP_MOUSE
|
||||||
| NC score | 0.068796 (rank : 113) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
FA8_MOUSE
|
||||||
| NC score | 0.068641 (rank : 114) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06194 | Gene names | F8, Cf8, F8c | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component). | |||||
|
FA8_HUMAN
|
||||||
| NC score | 0.068599 (rank : 115) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
PDGFD_MOUSE
|
||||||
| NC score | 0.068305 (rank : 116) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q925I7, Q3URF6, Q8K2L3, Q9D1L8 | Gene names | Pdgfd, Scdgfb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor chain precursor (PDGF D) (Spinal cord- derived growth factor B) (SCDGF-B). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.067004 (rank : 117) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.066841 (rank : 118) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
FA5_HUMAN
|
||||||
| NC score | 0.064774 (rank : 119) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
BMPER_HUMAN
|
||||||
| NC score | 0.064265 (rank : 120) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
NRCAM_HUMAN
|
||||||
| NC score | 0.063347 (rank : 121) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92823, O15051, O15179, Q9UHI3, Q9UHI4 | Gene names | NRCAM, KIAA0343 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal cell adhesion molecule precursor (Nr-CAM) (NgCAM-related cell adhesion molecule) (Ng-CAM-related) (hBravo). | |||||
|
BMPER_MOUSE
|
||||||
| NC score | 0.063113 (rank : 122) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
CNTN2_MOUSE
|
||||||
| NC score | 0.063035 (rank : 123) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61330, Q6NZJ4, Q7TSJ5 | Gene names | Cntn2, Tax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-2 precursor (Axonin-1) (Axonal glycoprotein TAG-1) (Transient axonal glycoprotein 1) (TAX-1). | |||||
|
CNTN2_HUMAN
|
||||||
| NC score | 0.062734 (rank : 124) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02246, P78432 | Gene names | CNTN2, TAG1, TAX1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-2 precursor (Axonin-1) (Axonal glycoprotein TAG-1) (Transient axonal glycoprotein 1) (TAX-1). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.062368 (rank : 125) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
DCC_HUMAN
|
||||||
| NC score | 0.061790 (rank : 126) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
CNTN3_MOUSE
|
||||||
| NC score | 0.061419 (rank : 127) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07409 | Gene names | Cntn3, Pang, Pcs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-3 precursor (Brain-derived immunoglobulin superfamily protein 1) (BIG-1) (Plasmacytoma-associated neuronal glycoprotein). | |||||
|
NFASC_MOUSE
|
||||||
| NC score | 0.059965 (rank : 128) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q810U3 | Gene names | Nfasc | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
NRCAM_MOUSE
|
||||||
| NC score | 0.059894 (rank : 129) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q810U4, Q80U33, Q8BLG8, Q8BX92, Q8BYJ8 | Gene names | Nrcam, Kiaa0343 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal cell adhesion molecule precursor (Nr-CAM) (NgCAM-related cell adhesion molecule) (Ng-CAM-related) (mBravo). | |||||
|
NEO1_MOUSE
|
||||||
| NC score | 0.059588 (rank : 130) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97798 | Gene names | Neo1, Ngn | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
DSCL1_HUMAN
|
||||||
| NC score | 0.059577 (rank : 131) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
CNTN4_HUMAN
|
||||||
| NC score | 0.059214 (rank : 132) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IWV2, Q8IX14, Q8TC35 | Gene names | CNTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-4 precursor (Brain-derived immunoglobulin superfamily protein 2) (BIG-2). | |||||
|
CNTN1_MOUSE
|
||||||
| NC score | 0.059016 (rank : 133) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P12960, Q6NXV7, Q8BR42, Q8C6A0 | Gene names | Cntn1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3). | |||||
|
DSCAM_HUMAN
|
||||||
| NC score | 0.058770 (rank : 134) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60469, O60468 | Gene names | DSCAM | |||
|
Domain Architecture |

|
|||||
| Description | Down syndrome cell adhesion molecule precursor (CHD2). | |||||
|
NEO1_HUMAN
|
||||||
| NC score | 0.058376 (rank : 135) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92859, O00340 | Gene names | NEO1, NGN | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
CNTN3_HUMAN
|
||||||
| NC score | 0.058148 (rank : 136) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P232, Q9H039 | Gene names | CNTN3, KIAA1496, PANG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-3 precursor (Brain-derived immunoglobulin superfamily protein 1) (BIG-1) (Plasmacytoma-associated neuronal glycoprotein). | |||||
|
DMBT1_MOUSE
|
||||||
| NC score | 0.058014 (rank : 137) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
L1CAM_HUMAN
|
||||||
| NC score | 0.057796 (rank : 138) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P32004, Q8TA87 | Gene names | L1CAM, CAML1, MIC5 | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule L1 precursor (N-CAM L1) (CD171 antigen). | |||||
|
CNTN1_HUMAN
|
||||||
| NC score | 0.057700 (rank : 139) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12860, Q12861, Q14030, Q8N466 | Gene names | CNTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3) (Glycoprotein gp135). | |||||
|
NFASC_HUMAN
|
||||||
| NC score | 0.057599 (rank : 140) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94856, Q5T2F0, Q5T2F1, Q5T2F2, Q5T2F3, Q5T2F4, Q5T2F5, Q5T2F6, Q5T2F7, Q5T2F9, Q5T2G0, Q5W9F8, Q68DH3, Q6ZQV6, Q7Z3K1, Q96HT1, Q96K50 | Gene names | NFASC, KIAA0756 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
CNTN6_HUMAN
|
||||||
| NC score | 0.056592 (rank : 141) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQ52 | Gene names | CNTN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (hNB-3). | |||||
|
CNTN4_MOUSE
|
||||||
| NC score | 0.056381 (rank : 142) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q69Z26, Q8BRT6, Q8CBD3 | Gene names | Cntn4, Kiaa3024 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-4 precursor (Brain-derived immunoglobulin superfamily protein 2) (BIG-2). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.056182 (rank : 143) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
CUBN_HUMAN
|
||||||
| NC score | 0.054752 (rank : 144) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
L1CAM_MOUSE
|
||||||
| NC score | 0.054361 (rank : 145) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P11627 | Gene names | L1cam, Caml1 | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule L1 precursor (N-CAM L1) (CD171 antigen). | |||||
|
CUBN_MOUSE
|
||||||
| NC score | 0.054074 (rank : 146) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.053935 (rank : 147) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
SDK1_MOUSE
|
||||||
| NC score | 0.053299 (rank : 148) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3UH53, Q3UFD1, Q6PAL2, Q6V3A4, Q8BMC2, Q8BZI1 | Gene names | Sdk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-1 precursor. | |||||
|
VWF_HUMAN
|
||||||
| NC score | 0.052833 (rank : 149) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
CNTN6_MOUSE
|
||||||
| NC score | 0.052764 (rank : 150) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JMB8, Q8C6X1 | Gene names | Cntn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (mNB-3). | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.052408 (rank : 151) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.052279 (rank : 152) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
LRP12_MOUSE
|
||||||
| NC score | 0.052210 (rank : 153) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
SDK2_MOUSE
|
||||||
| NC score | 0.051993 (rank : 154) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6V4S5, Q3TTK1, Q5U5W7, Q6ZPP2 | Gene names | Sdk2, Kiaa1514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-2 precursor. | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.051956 (rank : 155) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.051534 (rank : 156) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
SDK1_HUMAN
|
||||||
| NC score | 0.051485 (rank : 157) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z5N4, Q8TEN9, Q8TEP5, Q96N44 | Gene names | SDK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-1 precursor. | |||||
|
CSMD1_HUMAN
|
||||||
| NC score | 0.051244 (rank : 158) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
SDK2_HUMAN
|
||||||
| NC score | 0.050885 (rank : 159) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q58EX2, Q86VY3, Q9NTD2, Q9NVB3, Q9P214 | Gene names | SDK2, KIAA1514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein sidekick-2 precursor. | |||||
|
CSMD1_MOUSE
|
||||||
| NC score | 0.050879 (rank : 160) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
LRP12_HUMAN
|
||||||
| NC score | 0.050618 (rank : 161) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
TAF1C_MOUSE
|
||||||
| NC score | 0.019655 (rank : 162) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PDZ2, P97359, Q3U3B6, Q8BN54 | Gene names | Taf1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor, RNA polymerase I, subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 95 kDa) (TAFI95). | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.012382 (rank : 163) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.011738 (rank : 164) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
PRKDC_MOUSE
|
||||||
| NC score | 0.010528 (rank : 165) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
PC11Y_HUMAN
|
||||||
| NC score | 0.008620 (rank : 166) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZA8, Q70LR6, Q70LR8, Q70LS0, Q70LS1, Q70LS2, Q70LS3, Q70LS4, Q70LS5, Q8WY34, Q9BZA9, Q9H4E1 | Gene names | PCDH11Y, PCDH11, PCDH22, PCDHY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 Y-linked precursor (Protocadherin-11) (Protocadherin- 22) (Protocadherin on the Y chromosome) (PCDH-Y) (Protocadherin prostate cancer) (Protocadherin-PC). | |||||
|
ADA19_HUMAN
|
||||||
| NC score | 0.005045 (rank : 167) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H013, Q9BZL5, Q9UHP2 | Gene names | ADAM19, MLTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta) (Metalloprotease and disintegrin dentritic antigen marker) (MADDAM). | |||||
|
ZN546_HUMAN
|
||||||
| NC score | -0.001763 (rank : 168) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UE3 | Gene names | ZNF546, ZNF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 546. | |||||