Please be patient as the page loads
|
DHX57_MOUSE
|
||||||
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DHX57_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993141 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX36_MOUSE
|
||||||
| θ value | 1.15563e-172 (rank : 3) | NC score | 0.962172 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX36_HUMAN
|
||||||
| θ value | 1.50931e-172 (rank : 4) | NC score | 0.965085 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX29_MOUSE
|
||||||
| θ value | 3.72618e-163 (rank : 5) | NC score | 0.958497 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DHX29_HUMAN
|
||||||
| θ value | 4.55495e-161 (rank : 6) | NC score | 0.954149 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
DHX9_HUMAN
|
||||||
| θ value | 8.37844e-131 (rank : 7) | NC score | 0.954544 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
DHX9_MOUSE
|
||||||
| θ value | 9.26343e-130 (rank : 8) | NC score | 0.952725 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
DHX30_HUMAN
|
||||||
| θ value | 1.69605e-115 (rank : 9) | NC score | 0.936391 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_MOUSE
|
||||||
| θ value | 2.44908e-114 (rank : 10) | NC score | 0.935888 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX34_MOUSE
|
||||||
| θ value | 8.55514e-59 (rank : 11) | NC score | 0.895664 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX16_HUMAN
|
||||||
| θ value | 5.02714e-51 (rank : 12) | NC score | 0.834956 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
DHX8_HUMAN
|
||||||
| θ value | 1.61718e-49 (rank : 13) | NC score | 0.867564 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
DHX15_HUMAN
|
||||||
| θ value | 1.67352e-46 (rank : 14) | NC score | 0.878623 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43143, Q9NQT7 | Gene names | DHX15, DBP1, DDX15 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46). | |||||
|
DHX15_MOUSE
|
||||||
| θ value | 1.67352e-46 (rank : 15) | NC score | 0.878437 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35286, Q99L91 | Gene names | Dhx15, Ddx15, Deah9 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15). | |||||
|
PRP16_HUMAN
|
||||||
| θ value | 2.41655e-45 (rank : 16) | NC score | 0.865491 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
DHX34_HUMAN
|
||||||
| θ value | 3.15612e-45 (rank : 17) | NC score | 0.907324 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX35_HUMAN
|
||||||
| θ value | 4.55743e-44 (rank : 18) | NC score | 0.878300 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
DHX33_MOUSE
|
||||||
| θ value | 4.71619e-41 (rank : 19) | NC score | 0.867828 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80VY9, Q8BS50 | Gene names | Dhx33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX33_HUMAN
|
||||||
| θ value | 1.16164e-39 (rank : 20) | NC score | 0.863455 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H6R0, Q4G149, Q5CZ73, Q9H5M9 | Gene names | DHX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
YTDC2_HUMAN
|
||||||
| θ value | 3.4976e-36 (rank : 21) | NC score | 0.843440 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
DHX40_HUMAN
|
||||||
| θ value | 1.73584e-35 (rank : 22) | NC score | 0.861104 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
DHX40_MOUSE
|
||||||
| θ value | 2.26708e-35 (rank : 23) | NC score | 0.860268 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
DHX37_HUMAN
|
||||||
| θ value | 2.34606e-32 (rank : 24) | NC score | 0.872739 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
RBM22_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 25) | NC score | 0.088843 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 26) | NC score | 0.088843 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
MKRN2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 27) | NC score | 0.088957 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
SMBP2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 28) | NC score | 0.074412 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
SMBP2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 29) | NC score | 0.069688 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
PAR12_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 30) | NC score | 0.058427 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
MKRN4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.071968 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
MKRN2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.080088 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 33) | NC score | 0.028694 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
MKRN1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 34) | NC score | 0.074732 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
MKRN1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 35) | NC score | 0.075229 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
PAR12_HUMAN
|
||||||
| θ value | 0.279714 (rank : 36) | NC score | 0.053036 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.073034 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.007565 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
R113A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.053726 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
CO5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.012142 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01031 | Gene names | C5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
KIF6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.006325 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZMV9, Q2MDE3, Q2MDE4, Q5T8J6, Q6ZWE3, Q86T87, Q8WTV4 | Gene names | KIF6, C6orf102 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF6. | |||||
|
CREL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.010438 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
CREL1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.010294 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.034604 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
HIC2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | -0.002181 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96JB3, Q96KR3, Q9NSM9, Q9UPX9 | Gene names | HIC2, HRG22, KIAA1020, ZBTB30 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 2 protein (Hic-2) (Hic-3) (HIC1-related gene on chromosome 22) (Zinc finger and BTB domain-containing protein 30). | |||||
|
NUB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.037599 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5A7, O95422, Q8IX22, Q9BXR2 | Gene names | NUB1, NYREN18 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8 ultimate buster 1 (NY-REN-18 antigen). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.000316 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.028841 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.037032 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
EAF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.017450 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D4C5 | Gene names | Eaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
GP179_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.015441 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
HOOK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.005981 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.003115 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PI51C_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.010795 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
R113B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.042446 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IZP6, Q8WWF9, Q96QY9 | Gene names | RNF113B, RNF161, ZNF183L1 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113B (Zinc finger protein 183-like 1). | |||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.051468 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.050217 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
CN132_HUMAN
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.031781 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPU4 | Gene names | C14orf132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf132. | |||||
|
GRP75_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.008881 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.067224 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
NU107_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.032721 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57740 | Gene names | NUP107 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup107 (Nucleoporin Nup107) (107 kDa nucleoporin). | |||||
|
NUB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.035353 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54729, Q8K3U0 | Gene names | Nub1, Nyren18 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8 ultimate buster 1 (Protein BS4). | |||||
|
PKN3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | -0.001087 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P5Z2, Q9UM03 | Gene names | PKN3, PKNBETA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
TTP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.049226 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |
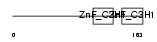
|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
TTP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.051083 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.053171 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
NUPL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.055204 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15504, Q49AE7, Q9BS49 | Gene names | NUPL2, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1) (hCG1) (NUP42 homolog). | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.051887 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.007003 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
APBB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.009428 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXJ1, O08642, Q3TPU0, Q8BNF4, Q8BSR9 | Gene names | Apbb1, Fe65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.010888 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.047700 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.049317 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.010671 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
GRP75_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.008343 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
NU107_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.029649 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BH74, Q99KH5 | Gene names | Nup107 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup107 (Nucleoporin Nup107) (107 kDa nucleoporin). | |||||
|
PLCB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.007776 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
U2AFL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.021365 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | -0.001793 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CPSF4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.057157 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
CPSF4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.056335 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
EP15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.003655 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.007665 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
PEO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.034314 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96RR1, Q6MZX2, Q96RR0 | Gene names | PEO1, C10orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein). | |||||
|
ZGPAT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.040792 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
ZGPAT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.038132 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | -0.001911 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
LRP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.002365 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
MRP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.018728 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
NEB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.011454 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
PEO1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.039049 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CIW5, Q8K1Z1 | Gene names | Peo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein homolog). | |||||
|
TCAL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.015883 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q921P9 | Gene names | Tceal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 1 (TCEA-like protein 1) (Transcription elongation factor S-II protein-like 1). | |||||
|
TR150_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.029098 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | -0.001777 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
MRP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.017448 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
NALP7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.006496 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NUPL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.034358 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CIC2, Q8BJX3, Q8BVP7, Q924S0 | Gene names | Nupl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1). | |||||
|
TE2IP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.012974 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYB0, Q8WYZ3, Q9NWR2 | Gene names | TERF2IP, RAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomeric repeat-binding factor 2-interacting protein 1 (TRF2- interacting telomeric protein Rap1) (hRap1). | |||||
|
TOP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.007857 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04750 | Gene names | Top1, Top-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
TR150_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.012143 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2W1, Q5VTK6 | Gene names | THRAP3, TRAP150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
TRPV5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.002613 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
U2AFL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.020180 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.018513 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
ZC3H8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.051883 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
ZC3H8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.052378 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.001550 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
F120C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.007212 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NX05 | Gene names | FAM120C, CXorf17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120C (Tumor antigen BJ-HCC-21). | |||||
|
GALT8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.001899 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NY28 | Gene names | GALNT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable polypeptide N-acetylgalactosaminyltransferase 8 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 8) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 8) (Polypeptide GalNAc transferase 8) (GalNAc-T8) (pp-GaNTase 8). | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.003628 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.001810 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.007436 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
NFM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.001780 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PKN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | -0.001924 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
SELS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.011238 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BCZ4, Q921S1, Q9DB55 | Gene names | Sels, H47, Vimp | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein S (VCP-interacting membrane protein) (Minor histocompatibility antigen H47). | |||||
|
U2AFM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.015272 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
AT8B4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.001402 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TF62, Q9H727 | Gene names | ATP8B4, KIAA1939 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IM (EC 3.6.3.1) (ATPase class I type 8B member 4). | |||||
|
CTGE6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.003001 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86UF2 | Gene names | CTAGE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-6 (cTAGE family member 6). | |||||
|
ERF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.010229 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62495, P46055, Q5M7Z7 | Gene names | ETF1, ERF1, RF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic peptide chain release factor subunit 1 (eRF1) (Eukaryotic release factor 1) (TB3-1) (Cl1 protein). | |||||
|
ERF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.010229 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BWY3, Q3TPZ6, Q91VH9 | Gene names | Etf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic peptide chain release factor subunit 1 (eRF1) (Eukaryotic release factor 1). | |||||
|
MBN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.018882 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R003 | Gene names | Mbnl3, Chcr, Mbxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein MCHCR). | |||||
|
MTP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.006478 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55157 | Gene names | MTTP, MTP | |||
|
Domain Architecture |

|
|||||
| Description | Microsomal triglyceride transfer protein large subunit precursor. | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.006125 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.002097 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
TISB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.028794 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |
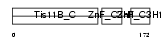
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
TTC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.002845 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.993141 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX36_HUMAN
|
||||||
| NC score | 0.965085 (rank : 3) | θ value | 1.50931e-172 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX36_MOUSE
|
||||||
| NC score | 0.962172 (rank : 4) | θ value | 1.15563e-172 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX29_MOUSE
|
||||||
| NC score | 0.958497 (rank : 5) | θ value | 3.72618e-163 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DHX9_HUMAN
|
||||||
| NC score | 0.954544 (rank : 6) | θ value | 8.37844e-131 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
DHX29_HUMAN
|
||||||
| NC score | 0.954149 (rank : 7) | θ value | 4.55495e-161 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
DHX9_MOUSE
|
||||||
| NC score | 0.952725 (rank : 8) | θ value | 9.26343e-130 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
DHX30_HUMAN
|
||||||
| NC score | 0.936391 (rank : 9) | θ value | 1.69605e-115 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_MOUSE
|
||||||
| NC score | 0.935888 (rank : 10) | θ value | 2.44908e-114 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX34_HUMAN
|
||||||
| NC score | 0.907324 (rank : 11) | θ value | 3.15612e-45 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX34_MOUSE
|
||||||
| NC score | 0.895664 (rank : 12) | θ value | 8.55514e-59 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX15_HUMAN
|
||||||
| NC score | 0.878623 (rank : 13) | θ value | 1.67352e-46 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43143, Q9NQT7 | Gene names | DHX15, DBP1, DDX15 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46). | |||||
|
DHX15_MOUSE
|
||||||
| NC score | 0.878437 (rank : 14) | θ value | 1.67352e-46 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35286, Q99L91 | Gene names | Dhx15, Ddx15, Deah9 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15). | |||||
|
DHX35_HUMAN
|
||||||
| NC score | 0.878300 (rank : 15) | θ value | 4.55743e-44 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
DHX37_HUMAN
|
||||||
| NC score | 0.872739 (rank : 16) | θ value | 2.34606e-32 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
DHX33_MOUSE
|
||||||
| NC score | 0.867828 (rank : 17) | θ value | 4.71619e-41 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80VY9, Q8BS50 | Gene names | Dhx33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX8_HUMAN
|
||||||
| NC score | 0.867564 (rank : 18) | θ value | 1.61718e-49 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
PRP16_HUMAN
|
||||||
| NC score | 0.865491 (rank : 19) | θ value | 2.41655e-45 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
DHX33_HUMAN
|
||||||
| NC score | 0.863455 (rank : 20) | θ value | 1.16164e-39 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H6R0, Q4G149, Q5CZ73, Q9H5M9 | Gene names | DHX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX40_HUMAN
|
||||||
| NC score | 0.861104 (rank : 21) | θ value | 1.73584e-35 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
DHX40_MOUSE
|
||||||
| NC score | 0.860268 (rank : 22) | θ value | 2.26708e-35 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
YTDC2_HUMAN
|
||||||
| NC score | 0.843440 (rank : 23) | θ value | 3.4976e-36 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
DHX16_HUMAN
|
||||||
| NC score | 0.834956 (rank : 24) | θ value | 5.02714e-51 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
MKRN2_MOUSE
|
||||||
| NC score | 0.088957 (rank : 25) | θ value | 0.0113563 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
RBM22_HUMAN
|
||||||
| NC score | 0.088843 (rank : 26) | θ value | 0.00175202 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| NC score | 0.088843 (rank : 27) | θ value | 0.00175202 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
MKRN2_HUMAN
|
||||||
| NC score | 0.080088 (rank : 28) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
MKRN1_MOUSE
|
||||||
| NC score | 0.075229 (rank : 29) | θ value | 0.21417 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
MKRN1_HUMAN
|
||||||
| NC score | 0.074732 (rank : 30) | θ value | 0.21417 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
SMBP2_HUMAN
|
||||||
| NC score | 0.074412 (rank : 31) | θ value | 0.0148317 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.073034 (rank : 32) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
MKRN4_HUMAN
|
||||||
| NC score | 0.071968 (rank : 33) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
SMBP2_MOUSE
|
||||||
| NC score | 0.069688 (rank : 34) | θ value | 0.0431538 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.067224 (rank : 35) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
PAR12_MOUSE
|
||||||
| NC score | 0.058427 (rank : 36) | θ value | 0.0563607 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
CPSF4_HUMAN
|
||||||
| NC score | 0.057157 (rank : 37) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
CPSF4_MOUSE
|
||||||
| NC score | 0.056335 (rank : 38) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
NUPL2_HUMAN
|
||||||
| NC score | 0.055204 (rank : 39) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15504, Q49AE7, Q9BS49 | Gene names | NUPL2, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1) (hCG1) (NUP42 homolog). | |||||
|
R113A_HUMAN
|
||||||
| NC score | 0.053726 (rank : 40) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.053171 (rank : 41) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
PAR12_HUMAN
|
||||||
| NC score | 0.053036 (rank : 42) | θ value | 0.279714 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
ZC3H8_MOUSE
|
||||||
| NC score | 0.052378 (rank : 43) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.051887 (rank : 44) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H8_HUMAN
|
||||||
| NC score | 0.051883 (rank : 45) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.051468 (rank : 46) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
TTP_MOUSE
|
||||||
| NC score | 0.051083 (rank : 47) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.050217 (rank : 48) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.049317 (rank : 49) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
TTP_HUMAN
|
||||||
| NC score | 0.049226 (rank : 50) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.047700 (rank : 51) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
R113B_HUMAN
|
||||||
| NC score | 0.042446 (rank : 52) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IZP6, Q8WWF9, Q96QY9 | Gene names | RNF113B, RNF161, ZNF183L1 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113B (Zinc finger protein 183-like 1). | |||||
|
ZGPAT_HUMAN
|
||||||
| NC score | 0.040792 (rank : 53) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
PEO1_MOUSE
|
||||||
| NC score | 0.039049 (rank : 54) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CIW5, Q8K1Z1 | Gene names | Peo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein homolog). | |||||
|
ZGPAT_MOUSE
|
||||||
| NC score | 0.038132 (rank : 55) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
NUB1_HUMAN
|
||||||
| NC score | 0.037599 (rank : 56) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5A7, O95422, Q8IX22, Q9BXR2 | Gene names | NUB1, NYREN18 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8 ultimate buster 1 (NY-REN-18 antigen). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.037032 (rank : 57) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
NUB1_MOUSE
|
||||||
| NC score | 0.035353 (rank : 58) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54729, Q8K3U0 | Gene names | Nub1, Nyren18 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8 ultimate buster 1 (Protein BS4). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.034604 (rank : 59) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
NUPL2_MOUSE
|
||||||
| NC score | 0.034358 (rank : 60) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CIC2, Q8BJX3, Q8BVP7, Q924S0 | Gene names | Nupl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1). | |||||
|
PEO1_HUMAN
|
||||||
| NC score | 0.034314 (rank : 61) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96RR1, Q6MZX2, Q96RR0 | Gene names | PEO1, C10orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein). | |||||
|
NU107_HUMAN
|
||||||
| NC score | 0.032721 (rank : 62) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57740 | Gene names | NUP107 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup107 (Nucleoporin Nup107) (107 kDa nucleoporin). | |||||
|
CN132_HUMAN
|
||||||
| NC score | 0.031781 (rank : 63) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPU4 | Gene names | C14orf132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf132. | |||||
|
NU107_MOUSE
|
||||||
| NC score | 0.029649 (rank : 64) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BH74, Q99KH5 | Gene names | Nup107 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup107 (Nucleoporin Nup107) (107 kDa nucleoporin). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.029098 (rank : 65) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.028841 (rank : 66) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
TISB_MOUSE
|
||||||
| NC score | 0.028794 (rank : 67) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.028694 (rank : 68) | θ value | 0.163984 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
U2AFL_MOUSE
|
||||||
| NC score | 0.021365 (rank : 69) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
U2AFL_HUMAN
|
||||||
| NC score | 0.020180 (rank : 70) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
MBN3_MOUSE
|
||||||
| NC score | 0.018882 (rank : 71) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R003 | Gene names | Mbnl3, Chcr, Mbxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein MCHCR). | |||||
|
MRP_MOUSE
|
||||||
| NC score | 0.018728 (rank : 72) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.018513 (rank : 73) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
EAF1_MOUSE
|
||||||
| NC score | 0.017450 (rank : 74) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D4C5 | Gene names | Eaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
MRP_HUMAN
|
||||||
| NC score | 0.017448 (rank : 75) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
TCAL1_MOUSE
|
||||||
| NC score | 0.015883 (rank : 76) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q921P9 | Gene names | Tceal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 1 (TCEA-like protein 1) (Transcription elongation factor S-II protein-like 1). | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.015441 (rank : 77) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
U2AFM_MOUSE
|
||||||
| NC score | 0.015272 (rank : 78) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
TE2IP_HUMAN
|
||||||
| NC score | 0.012974 (rank : 79) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYB0, Q8WYZ3, Q9NWR2 | Gene names | TERF2IP, RAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomeric repeat-binding factor 2-interacting protein 1 (TRF2- interacting telomeric protein Rap1) (hRap1). | |||||
|
TR150_HUMAN
|
||||||
| NC score | 0.012143 (rank : 80) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2W1, Q5VTK6 | Gene names | THRAP3, TRAP150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
CO5_HUMAN
|
||||||
| NC score | 0.012142 (rank : 81) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01031 | Gene names | C5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.011454 (rank : 82) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
SELS_MOUSE
|
||||||
| NC score | 0.011238 (rank : 83) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BCZ4, Q921S1, Q9DB55 | Gene names | Sels, H47, Vimp | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein S (VCP-interacting membrane protein) (Minor histocompatibility antigen H47). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.010888 (rank : 84) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
PI51C_HUMAN
|
||||||
| NC score | 0.010795 (rank : 85) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.010671 (rank : 86) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
CREL1_HUMAN
|
||||||
| NC score | 0.010438 (rank : 87) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
CREL1_MOUSE
|
||||||
| NC score | 0.010294 (rank : 88) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
ERF1_HUMAN
|
||||||
| NC score | 0.010229 (rank : 89) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62495, P46055, Q5M7Z7 | Gene names | ETF1, ERF1, RF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic peptide chain release factor subunit 1 (eRF1) (Eukaryotic release factor 1) (TB3-1) (Cl1 protein). | |||||
|
ERF1_MOUSE
|
||||||
| NC score | 0.010229 (rank : 90) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BWY3, Q3TPZ6, Q91VH9 | Gene names | Etf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic peptide chain release factor subunit 1 (eRF1) (Eukaryotic release factor 1). | |||||
|
APBB1_MOUSE
|
||||||
| NC score | 0.009428 (rank : 91) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXJ1, O08642, Q3TPU0, Q8BNF4, Q8BSR9 | Gene names | Apbb1, Fe65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
GRP75_MOUSE
|
||||||
| NC score | 0.008881 (rank : 92) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
GRP75_HUMAN
|
||||||
| NC score | 0.008343 (rank : 93) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
TOP1_MOUSE
|
||||||
| NC score | 0.007857 (rank : 94) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04750 | Gene names | Top1, Top-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
PLCB1_MOUSE
|
||||||
| NC score | 0.007776 (rank : 95) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.007665 (rank : 96) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.007565 (rank : 97) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.007436 (rank : 98) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
F120C_HUMAN
|
||||||
| NC score | 0.007212 (rank : 99) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NX05 | Gene names | FAM120C, CXorf17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120C (Tumor antigen BJ-HCC-21). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.007003 (rank : 100) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
NALP7_HUMAN
|
||||||
| NC score | 0.006496 (rank : 101) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
MTP_HUMAN
|
||||||
| NC score | 0.006478 (rank : 102) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55157 | Gene names | MTTP, MTP | |||
|
Domain Architecture |

|
|||||
| Description | Microsomal triglyceride transfer protein large subunit precursor. | |||||
|
KIF6_HUMAN
|
||||||
| NC score | 0.006325 (rank : 103) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZMV9, Q2MDE3, Q2MDE4, Q5T8J6, Q6ZWE3, Q86T87, Q8WTV4 | Gene names | KIF6, C6orf102 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF6. | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.006125 (rank : 104) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
HOOK1_MOUSE
|
||||||
| NC score | 0.005981 (rank : 105) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.003655 (rank : 106) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.003628 (rank : 107) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.003115 (rank : 108) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CTGE6_HUMAN
|
||||||
| NC score | 0.003001 (rank : 109) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86UF2 | Gene names | CTAGE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-6 (cTAGE family member 6). | |||||
|
TTC3_HUMAN
|
||||||
| NC score | 0.002845 (rank : 110) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
TRPV5_HUMAN
|
||||||
| NC score | 0.002613 (rank : 111) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.002365 (rank : 112) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.002097 (rank : 113) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
GALT8_HUMAN
|
||||||
| NC score | 0.001899 (rank : 114) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NY28 | Gene names | GALNT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable polypeptide N-acetylgalactosaminyltransferase 8 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 8) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 8) (Polypeptide GalNAc transferase 8) (GalNAc-T8) (pp-GaNTase 8). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.001810 (rank : 115) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.001780 (rank : 116) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.001550 (rank : 117) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
AT8B4_HUMAN
|
||||||
| NC score | 0.001402 (rank : 118) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TF62, Q9H727 | Gene names | ATP8B4, KIAA1939 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IM (EC 3.6.3.1) (ATPase class I type 8B member 4). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.000316 (rank : 119) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
PKN3_HUMAN
|
||||||
| NC score | -0.001087 (rank : 120) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P5Z2, Q9UM03 | Gene names | PKN3, PKNBETA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | -0.001777 (rank : 121) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | -0.001793 (rank : 122) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | -0.001911 (rank : 123) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
PKN3_MOUSE
|
||||||
| NC score | -0.001924 (rank : 124) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
HIC2_HUMAN
|
||||||
| NC score | -0.002181 (rank : 125) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96JB3, Q96KR3, Q9NSM9, Q9UPX9 | Gene names | HIC2, HRG22, KIAA1020, ZBTB30 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 2 protein (Hic-2) (Hic-3) (HIC1-related gene on chromosome 22) (Zinc finger and BTB domain-containing protein 30). | |||||