Please be patient as the page loads
|
ZC3H3_MOUSE
|
||||||
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.941185 (rank : 2) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 188 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
CPSF4_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 3) | NC score | 0.641687 (rank : 3) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
CPSF4_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 4) | NC score | 0.584630 (rank : 4) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 5) | NC score | 0.387822 (rank : 7) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
MBN3_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 6) | NC score | 0.326488 (rank : 11) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NUK0, Q5JXN8, Q5JXN9, Q5JXP4, Q6UDQ1, Q8TAD9, Q8TAF4, Q9H0Z7, Q9UF37 | Gene names | MBNL3, CHCR, MBLX39, MBXL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein HCHCR). | |||||
|
MBN3_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 7) | NC score | 0.319947 (rank : 14) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R003 | Gene names | Mbnl3, Chcr, Mbxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein MCHCR). | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 8) | NC score | 0.374889 (rank : 8) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 9) | NC score | 0.358028 (rank : 10) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
MKRN2_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 10) | NC score | 0.322224 (rank : 12) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
MKRN2_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 11) | NC score | 0.321875 (rank : 13) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 12) | NC score | 0.360749 (rank : 9) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
ZC3H8_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 13) | NC score | 0.403887 (rank : 6) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
ZC3H8_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 14) | NC score | 0.405035 (rank : 5) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
MBNL_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 15) | NC score | 0.293010 (rank : 17) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NR56, O43311, O43797 | Gene names | MBNL1, EXP, KIAA0428, MBNL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
MBNL_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 16) | NC score | 0.295579 (rank : 15) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JKP5 | Gene names | Mbnl1, Exp, Mbnl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
MKRN1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 17) | NC score | 0.291546 (rank : 18) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
MKRN1_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 18) | NC score | 0.294862 (rank : 16) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 19) | NC score | 0.118915 (rank : 37) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MKRN4_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 20) | NC score | 0.250514 (rank : 21) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
PHLB1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 21) | NC score | 0.054719 (rank : 59) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 22) | NC score | 0.071521 (rank : 46) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
TTP_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 23) | NC score | 0.270983 (rank : 19) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |
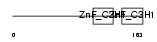
|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
MNAB_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 24) | NC score | 0.119139 (rank : 36) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P0C090 | Gene names | Mnab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 25) | NC score | 0.037131 (rank : 90) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 26) | NC score | 0.037764 (rank : 89) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
TISD_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 27) | NC score | 0.236332 (rank : 23) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |
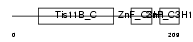
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
ATRIP_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 28) | NC score | 0.058707 (rank : 54) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXE1, Q8NHQ2, Q8WUG7, Q96CL3, Q9HA30 | Gene names | ATRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATR-interacting protein (ATM and Rad3-related-interacting protein). | |||||
|
LEUK_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 29) | NC score | 0.083638 (rank : 45) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
ADA17_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 30) | NC score | 0.052414 (rank : 63) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78536, O60226 | Gene names | ADAM17, CSVP, TACE | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (Snake venom-like protease) (CD156b antigen). | |||||
|
TISD_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 31) | NC score | 0.242135 (rank : 22) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |
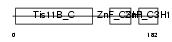
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 32) | NC score | 0.065213 (rank : 47) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
TTP_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 33) | NC score | 0.266786 (rank : 20) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
PAR12_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 34) | NC score | 0.132491 (rank : 32) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
RC3H1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 35) | NC score | 0.124712 (rank : 33) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
FOXC2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.029302 (rank : 106) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |
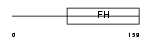
|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
TISB_HUMAN
|
||||||
| θ value | 0.125558 (rank : 37) | NC score | 0.234260 (rank : 24) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |
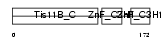
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
TISB_MOUSE
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.232802 (rank : 25) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |
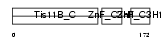
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
ADA17_MOUSE
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.047516 (rank : 73) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0F8, O88726, Q9R1U4, Q9Z0K3 | Gene names | Adam17, Tace | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (CD156b antigen). | |||||
|
PARPT_HUMAN
|
||||||
| θ value | 0.163984 (rank : 40) | NC score | 0.102876 (rank : 39) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
PARPT_MOUSE
|
||||||
| θ value | 0.163984 (rank : 41) | NC score | 0.102668 (rank : 40) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
ULK1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 42) | NC score | 0.007449 (rank : 179) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
DDX24_MOUSE
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.020309 (rank : 142) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 0.21417 (rank : 44) | NC score | 0.086887 (rank : 44) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
ZC3H5_MOUSE
|
||||||
| θ value | 0.21417 (rank : 45) | NC score | 0.166530 (rank : 28) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.279714 (rank : 46) | NC score | 0.050521 (rank : 66) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.097473 (rank : 43) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ZC11A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.172903 (rank : 26) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75152, Q6AHY4, Q6AHY9, Q6AW79, Q6AWA1, Q6PJK4, Q86XZ7 | Gene names | ZC3H11A, KIAA0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.046730 (rank : 75) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
UBP32_HUMAN
|
||||||
| θ value | 0.365318 (rank : 50) | NC score | 0.023465 (rank : 134) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
ZC3H5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 51) | NC score | 0.157446 (rank : 30) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9C0B0 | Gene names | ZC3H5, KIAA1753, ZC3HDC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.024450 (rank : 128) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
FOXJ3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.028169 (rank : 111) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
PAR12_MOUSE
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.123420 (rank : 35) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
ZC11A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.156475 (rank : 31) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
CT075_HUMAN
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.027284 (rank : 116) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WUT4, Q5JWV6, Q9H419 | Gene names | C20orf75 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf75 precursor. | |||||
|
CTTB2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 57) | NC score | 0.020047 (rank : 143) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 58) | NC score | 0.021101 (rank : 139) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.045466 (rank : 76) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
PIGQ_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.042949 (rank : 79) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BRB3, O14927, Q96G00, Q96S22, Q9UJH4 | Gene names | PIGQ, GPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1). | |||||
|
RC3H1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.124501 (rank : 34) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q4VGL6, Q69Z31 | Gene names | Rc3h1, Gm551, Kiaa2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1) (Sanroque protein). | |||||
|
RP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.032443 (rank : 103) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56715 | Gene names | RP1, ORP1 | |||
|
Domain Architecture |
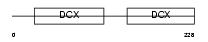
|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein) (Retinitis pigmentosa 1 protein). | |||||
|
WDR46_MOUSE
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.040100 (rank : 84) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z0H1 | Gene names | Wdr46, Bing4 | |||
|
Domain Architecture |
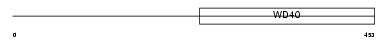
|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.065125 (rank : 48) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
GNRP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.019161 (rank : 144) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
PZRN3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 66) | NC score | 0.020382 (rank : 141) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
RBM22_HUMAN
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.099895 (rank : 41) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.099895 (rank : 42) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
CEP41_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.038981 (rank : 87) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYV8, Q7Z496, Q86TM1, Q8NFU8, Q9H6A3, Q9NPV3 | Gene names | TSGA14, CEP41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.050217 (rank : 70) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
GEMI5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.023936 (rank : 130) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BX17 | Gene names | Gemin5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.032451 (rank : 102) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 73) | NC score | 0.024370 (rank : 129) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.025666 (rank : 125) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TTLL5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 75) | NC score | 0.026682 (rank : 118) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
ZN297_MOUSE
|
||||||
| θ value | 1.06291 (rank : 76) | NC score | 0.005757 (rank : 182) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z0G7 | Gene names | Znf297, Bing1, Zfp297 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 297 (BING1 protein). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.008910 (rank : 175) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
ANXA7_MOUSE
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.014907 (rank : 157) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07076 | Gene names | Anxa7, Anx7 | |||
|
Domain Architecture |
|
|||||
| Description | Annexin A7 (Annexin VII) (Synexin). | |||||
|
CEND_HUMAN
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | 0.059122 (rank : 53) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N111, Q9NYM6 | Gene names | CEND1, BM88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle exit and neuronal differentiation protein 1 (BM88 antigen). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.056214 (rank : 57) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
ELF2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.016972 (rank : 150) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ILDR1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.034707 (rank : 95) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86SU0, Q6ZP61, Q7Z578 | Gene names | ILDR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin-like domain-containing receptor 1 precursor. | |||||
|
K1849_MOUSE
|
||||||
| θ value | 1.38821 (rank : 83) | NC score | 0.026125 (rank : 123) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
MNAB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.105036 (rank : 38) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HBD1, Q5JPD7, Q86ST6, Q8N3D6, Q96F27, Q9H5J2, Q9HBD2, Q9NWN9, Q9NXE1 | Gene names | MNAB, RNF164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein (RING finger protein 164). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 85) | NC score | 0.033585 (rank : 99) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
CP007_MOUSE
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.034864 (rank : 94) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.056280 (rank : 56) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.027901 (rank : 113) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.026275 (rank : 121) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.009527 (rank : 173) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 91) | NC score | 0.023003 (rank : 135) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 1.81305 (rank : 92) | NC score | 0.028724 (rank : 109) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 1.81305 (rank : 93) | NC score | 0.052702 (rank : 62) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
ZN330_HUMAN
|
||||||
| θ value | 1.81305 (rank : 94) | NC score | 0.061672 (rank : 50) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3S2 | Gene names | ZNF330, NOA36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 330 (Nucleolar cysteine-rich protein) (Nucleolar autoantigen 36). | |||||
|
ZN330_MOUSE
|
||||||
| θ value | 1.81305 (rank : 95) | NC score | 0.061608 (rank : 51) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q922H9, Q8C389, Q8K2M4 | Gene names | Znf330, Noa36, Zfp330 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 330 (Nucleolar cysteine-rich protein) (Nucleolar autoantigen 36). | |||||
|
ZN509_MOUSE
|
||||||
| θ value | 1.81305 (rank : 96) | NC score | 0.000499 (rank : 193) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
BICD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.010804 (rank : 170) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.026166 (rank : 122) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
EVI2A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.036363 (rank : 92) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20934 | Gene names | Evi2a, Evi-2, Evi-2a, Evi2 | |||
|
Domain Architecture |
|
|||||
| Description | EVI2A protein precursor (Ecotropic viral integration site 2A protein). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.050246 (rank : 68) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
MAVS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.053550 (rank : 61) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 102) | NC score | 0.010587 (rank : 171) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 103) | NC score | 0.056698 (rank : 55) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PCD12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | 0.004073 (rank : 187) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NPG4, Q6UXB6, Q96KB8, Q9H7Y6, Q9H8E0 | Gene names | PCDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
SPIT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.027639 (rank : 114) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.012474 (rank : 166) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.025274 (rank : 126) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CT174_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.052233 (rank : 65) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.013486 (rank : 162) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
FOXC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.023668 (rank : 132) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
FOXJ3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.024962 (rank : 127) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BUR3, Q3UTD8 | Gene names | Foxj3, Kiaa1041 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein J3. | |||||
|
MEF2D_HUMAN
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.023537 (rank : 133) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14814, Q14815 | Gene names | MEF2D | |||
|
Domain Architecture |
|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
PITM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.014518 (rank : 159) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.034251 (rank : 97) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.027620 (rank : 115) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | 0.026564 (rank : 119) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.021962 (rank : 137) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TMM29_HUMAN
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | 0.041525 (rank : 81) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NDB6, Q5JPL2, Q5JPL3, Q5JPL4, Q5JPL5, Q5JPL6, Q68D17, Q6NVY3, Q9UI49 | Gene names | TMEM29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 29. | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.029490 (rank : 105) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.032931 (rank : 100) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.049080 (rank : 71) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.048605 (rank : 72) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.034366 (rank : 96) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
ELOA2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.023852 (rank : 131) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
GALT4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.005793 (rank : 181) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08832, Q3U681 | Gene names | Galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.028068 (rank : 112) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | 0.026296 (rank : 120) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 128) | NC score | 0.054987 (rank : 58) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 129) | NC score | 0.043512 (rank : 78) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 130) | NC score | 0.026740 (rank : 117) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
STAM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 131) | NC score | 0.012456 (rank : 167) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 132) | NC score | 0.005166 (rank : 185) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ZFP28_HUMAN
|
||||||
| θ value | 4.03905 (rank : 133) | NC score | -0.001710 (rank : 194) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NHY6, Q9BY30, Q9P2B6 | Gene names | ZFP28, KIAA1431 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 28 homolog (Zfp-28) (Krueppel-like zinc finger factor X6). | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.001841 (rank : 190) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.008831 (rank : 176) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
DOK7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.041669 (rank : 80) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.050229 (rank : 69) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
FOXM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.021554 (rank : 138) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
GASP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.015600 (rank : 154) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.008825 (rank : 177) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.022003 (rank : 136) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.031163 (rank : 104) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.036859 (rank : 91) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MRIP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 144) | NC score | 0.013838 (rank : 160) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 145) | NC score | 0.029208 (rank : 107) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NEO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 146) | NC score | 0.005220 (rank : 184) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97798 | Gene names | Neo1, Ngn | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
ROBO3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 147) | NC score | 0.006799 (rank : 180) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 148) | NC score | 0.028419 (rank : 110) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 149) | NC score | 0.011745 (rank : 169) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
TM108_MOUSE
|
||||||
| θ value | 5.27518 (rank : 150) | NC score | 0.033697 (rank : 98) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
TTC28_MOUSE
|
||||||
| θ value | 5.27518 (rank : 151) | NC score | 0.018259 (rank : 145) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 5.27518 (rank : 152) | NC score | 0.015349 (rank : 155) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
ALX4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.001352 (rank : 191) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35137 | Gene names | Alx4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein aristaless-like 4 (ALX-4). | |||||
|
DLGP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.012551 (rank : 164) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14490, O14489, P78335 | Gene names | DLGAP1, DAP1, GKAP | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 1 (DAP-1) (Guanylate kinase-associated protein) (hGKAP) (SAP90/PSD-95-associated protein 1) (SAPAP1) (PSD- 95/SAP90-binding protein 1). | |||||
|
FRAT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.015051 (rank : 156) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70339 | Gene names | Frat1 | |||
|
Domain Architecture |
|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
K1543_MOUSE
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.029082 (rank : 108) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
LAP2A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.013677 (rank : 161) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
|
LDB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.018089 (rank : 146) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.047170 (rank : 74) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NEO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | 0.004507 (rank : 186) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92859, O00340 | Gene names | NEO1, NGN | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
NIPA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.020489 (rank : 140) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80YV2, Q3TJE6, Q80Z11, Q8BTW5, Q8CI56, Q8R3U7 | Gene names | Zc3hc1, Nipa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear-interacting partner of ALK (Nuclear-interacting partner of anaplastic lymphoma kinase) (mNIPA) (Zinc finger C3HC-type protein 1). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 162) | NC score | 0.039792 (rank : 85) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 6.88961 (rank : 163) | NC score | 0.008268 (rank : 178) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 164) | NC score | 0.038033 (rank : 88) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 165) | NC score | 0.035166 (rank : 93) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RMI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 166) | NC score | 0.017852 (rank : 148) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D4G9, Q8C560, Q8CI20 | Gene names | Rmi1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RMI1 homolog. | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 6.88961 (rank : 167) | NC score | 0.017431 (rank : 149) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 168) | NC score | 0.026058 (rank : 124) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
UBP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 169) | NC score | 0.009501 (rank : 174) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UTY_HUMAN
|
||||||
| θ value | 6.88961 (rank : 170) | NC score | 0.012302 (rank : 168) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.032901 (rank : 101) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.041076 (rank : 82) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.040156 (rank : 83) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CT075_MOUSE
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.014568 (rank : 158) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59383 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf75 homolog precursor. | |||||
|
DCX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.016561 (rank : 151) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
FBN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.002037 (rank : 189) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
GRAM3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.013255 (rank : 163) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PEM6 | Gene names | Gramd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRAM domain-containing protein 3. | |||||
|
HIPK3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | 0.000626 (rank : 192) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ERH7, O88906, Q9QZR2 | Gene names | Hipk3, Fist3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeodomain-interacting protein kinase 3 (EC 2.7.11.1) (Fas- interacting serine/threonine-protein kinase) (FIST) (Nuclear body- associated kinase 3) (Nbak3) (Androgen receptor-interacting nuclear protein kinase) (ANPK). | |||||
|
MAST3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 179) | NC score | 0.002771 (rank : 188) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
MEF2D_MOUSE
|
||||||
| θ value | 8.99809 (rank : 180) | NC score | 0.016048 (rank : 152) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q63943, Q63944 | Gene names | Mef2d | |||
|
Domain Architecture |
|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
MPDZ_MOUSE
|
||||||
| θ value | 8.99809 (rank : 181) | NC score | 0.012474 (rank : 165) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 182) | NC score | 0.039408 (rank : 86) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PRGC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 183) | NC score | 0.018047 (rank : 147) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 184) | NC score | 0.015619 (rank : 153) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 185) | NC score | 0.064807 (rank : 49) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 186) | NC score | 0.044287 (rank : 77) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TLE3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 187) | NC score | 0.005295 (rank : 183) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q08122, Q923A4 | Gene names | Tle3, Esg | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG) (Grg-3). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 188) | NC score | 0.010141 (rank : 172) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
BSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.059768 (rank : 52) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.169194 (rank : 27) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.165557 (rank : 29) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.054310 (rank : 60) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.052370 (rank : 64) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.050474 (rank : 67) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 188 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.941185 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
CPSF4_HUMAN
|
||||||
| NC score | 0.641687 (rank : 3) | θ value | 2.06002e-20 (rank : 3) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
CPSF4_MOUSE
|
||||||
| NC score | 0.584630 (rank : 4) | θ value | 3.52202e-12 (rank : 4) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
ZC3H8_HUMAN
|
||||||
| NC score | 0.405035 (rank : 5) | θ value | 5.44631e-05 (rank : 14) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
ZC3H8_MOUSE
|
||||||
| NC score | 0.403887 (rank : 6) | θ value | 3.19293e-05 (rank : 13) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.387822 (rank : 7) | θ value | 4.45536e-07 (rank : 5) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.374889 (rank : 8) | θ value | 8.40245e-06 (rank : 8) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.360749 (rank : 9) | θ value | 3.19293e-05 (rank : 12) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.358028 (rank : 10) | θ value | 1.09739e-05 (rank : 9) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
MBN3_HUMAN
|
||||||
| NC score | 0.326488 (rank : 11) | θ value | 6.43352e-06 (rank : 6) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NUK0, Q5JXN8, Q5JXN9, Q5JXP4, Q6UDQ1, Q8TAD9, Q8TAF4, Q9H0Z7, Q9UF37 | Gene names | MBNL3, CHCR, MBLX39, MBXL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein HCHCR). | |||||
|
MKRN2_HUMAN
|
||||||
| NC score | 0.322224 (rank : 12) | θ value | 1.87187e-05 (rank : 10) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
MKRN2_MOUSE
|
||||||
| NC score | 0.321875 (rank : 13) | θ value | 2.44474e-05 (rank : 11) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
MBN3_MOUSE
|
||||||
| NC score | 0.319947 (rank : 14) | θ value | 8.40245e-06 (rank : 7) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R003 | Gene names | Mbnl3, Chcr, Mbxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein MCHCR). | |||||
|
MBNL_MOUSE
|
||||||
| NC score | 0.295579 (rank : 15) | θ value | 7.1131e-05 (rank : 16) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JKP5 | Gene names | Mbnl1, Exp, Mbnl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
MKRN1_MOUSE
|
||||||
| NC score | 0.294862 (rank : 16) | θ value | 0.00020696 (rank : 18) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
MBNL_HUMAN
|
||||||
| NC score | 0.293010 (rank : 17) | θ value | 7.1131e-05 (rank : 15) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NR56, O43311, O43797 | Gene names | MBNL1, EXP, KIAA0428, MBNL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
MKRN1_HUMAN
|
||||||
| NC score | 0.291546 (rank : 18) | θ value | 0.00020696 (rank : 17) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
TTP_HUMAN
|
||||||
| NC score | 0.270983 (rank : 19) | θ value | 0.00869519 (rank : 23) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
TTP_MOUSE
|
||||||
| NC score | 0.266786 (rank : 20) | θ value | 0.0736092 (rank : 33) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
MKRN4_HUMAN
|
||||||
| NC score | 0.250514 (rank : 21) | θ value | 0.00509761 (rank : 20) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
TISD_MOUSE
|
||||||
| NC score | 0.242135 (rank : 22) | θ value | 0.0563607 (rank : 31) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
TISD_HUMAN
|
||||||
| NC score | 0.236332 (rank : 23) | θ value | 0.0252991 (rank : 27) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
TISB_HUMAN
|
||||||
| NC score | 0.234260 (rank : 24) | θ value | 0.125558 (rank : 37) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
TISB_MOUSE
|
||||||
| NC score | 0.232802 (rank : 25) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
ZC11A_HUMAN
|
||||||
| NC score | 0.172903 (rank : 26) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75152, Q6AHY4, Q6AHY9, Q6AW79, Q6AWA1, Q6PJK4, Q86XZ7 | Gene names | ZC3H11A, KIAA0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.169194 (rank : 27) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
ZC3H5_MOUSE
|
||||||
| NC score | 0.166530 (rank : 28) | θ value | 0.21417 (rank : 45) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.165557 (rank : 29) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
ZC3H5_HUMAN
|
||||||
| NC score | 0.157446 (rank : 30) | θ value | 0.365318 (rank : 51) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9C0B0 | Gene names | ZC3H5, KIAA1753, ZC3HDC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
ZC11A_MOUSE
|
||||||
| NC score | 0.156475 (rank : 31) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
PAR12_HUMAN
|
||||||
| NC score | 0.132491 (rank : 32) | θ value | 0.0961366 (rank : 34) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
RC3H1_HUMAN
|
||||||
| NC score | 0.124712 (rank : 33) | θ value | 0.0961366 (rank : 35) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
RC3H1_MOUSE
|
||||||
| NC score | 0.124501 (rank : 34) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q4VGL6, Q69Z31 | Gene names | Rc3h1, Gm551, Kiaa2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1) (Sanroque protein). | |||||
|
PAR12_MOUSE
|
||||||
| NC score | 0.123420 (rank : 35) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
MNAB_MOUSE
|
||||||
| NC score | 0.119139 (rank : 36) | θ value | 0.0148317 (rank : 24) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P0C090 | Gene names | Mnab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.118915 (rank : 37) | θ value | 0.000602161 (rank : 19) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MNAB_HUMAN
|
||||||
| NC score | 0.105036 (rank : 38) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HBD1, Q5JPD7, Q86ST6, Q8N3D6, Q96F27, Q9H5J2, Q9HBD2, Q9NWN9, Q9NXE1 | Gene names | MNAB, RNF164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein (RING finger protein 164). | |||||
|
PARPT_HUMAN
|
||||||
| NC score | 0.102876 (rank : 39) | θ value | 0.163984 (rank : 40) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
PARPT_MOUSE
|
||||||
| NC score | 0.102668 (rank : 40) | θ value | 0.163984 (rank : 41) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
RBM22_HUMAN
|
||||||
| NC score | 0.099895 (rank : 41) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| NC score | 0.099895 (rank : 42) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.097473 (rank : 43) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.086887 (rank : 44) | θ value | 0.21417 (rank : 44) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
LEUK_HUMAN
|
||||||
| NC score | 0.083638 (rank : 45) | θ value | 0.0431538 (rank : 29) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.071521 (rank : 46) | θ value | 0.00665767 (rank : 22) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.065213 (rank : 47) | θ value | 0.0736092 (rank : 32) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.065125 (rank : 48) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.064807 (rank : 49) | θ value | 8.99809 (rank : 185) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
ZN330_HUMAN
|
||||||
| NC score | 0.061672 (rank : 50) | θ value | 1.81305 (rank : 94) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3S2 | Gene names | ZNF330, NOA36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 330 (Nucleolar cysteine-rich protein) (Nucleolar autoantigen 36). | |||||
|
ZN330_MOUSE
|
||||||
| NC score | 0.061608 (rank : 51) | θ value | 1.81305 (rank : 95) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q922H9, Q8C389, Q8K2M4 | Gene names | Znf330, Noa36, Zfp330 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 330 (Nucleolar cysteine-rich protein) (Nucleolar autoantigen 36). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.059768 (rank : 52) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CEND_HUMAN
|
||||||
| NC score | 0.059122 (rank : 53) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N111, Q9NYM6 | Gene names | CEND1, BM88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle exit and neuronal differentiation protein 1 (BM88 antigen). | |||||
|
ATRIP_HUMAN
|
||||||
| NC score | 0.058707 (rank : 54) | θ value | 0.0431538 (rank : 28) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXE1, Q8NHQ2, Q8WUG7, Q96CL3, Q9HA30 | Gene names | ATRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATR-interacting protein (ATM and Rad3-related-interacting protein). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.056698 (rank : 55) | θ value | 2.36792 (rank : 103) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.056280 (rank : 56) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.056214 (rank : 57) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.054987 (rank : 58) | θ value | 4.03905 (rank : 128) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PHLB1_MOUSE
|
||||||
| NC score | 0.054719 (rank : 59) | θ value | 0.00509761 (rank : 21) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.054310 (rank : 60) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MAVS_HUMAN
|
||||||
| NC score | 0.053550 (rank : 61) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.052702 (rank : 62) | θ value | 1.81305 (rank : 93) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
ADA17_HUMAN
|
||||||
| NC score | 0.052414 (rank : 63) | θ value | 0.0563607 (rank : 30) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78536, O60226 | Gene names | ADAM17, CSVP, TACE | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (Snake venom-like protease) (CD156b antigen). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.052370 (rank : 64) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CT174_HUMAN
|
||||||
| NC score | 0.052233 (rank : 65) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.050521 (rank : 66) | θ value | 0.279714 (rank : 46) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.050474 (rank : 67) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.050246 (rank : 68) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.050229 (rank : 69) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.050217 (rank : 70) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.049080 (rank : 71) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.048605 (rank : 72) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
ADA17_MOUSE
|
||||||
| NC score | 0.047516 (rank : 73) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0F8, O88726, Q9R1U4, Q9Z0K3 | Gene names | Adam17, Tace | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (CD156b antigen). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.047170 (rank : 74) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.046730 (rank : 75) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.045466 (rank : 76) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.044287 (rank : 77) | θ value | 8.99809 (rank : 186) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.043512 (rank : 78) | θ value | 4.03905 (rank : 129) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
PIGQ_HUMAN
|
||||||
| NC score | 0.042949 (rank : 79) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BRB3, O14927, Q96G00, Q96S22, Q9UJH4 | Gene names | PIGQ, GPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1). | |||||
|
DOK7_MOUSE
|
||||||
| NC score | 0.041669 (rank : 80) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
TMM29_HUMAN
|
||||||
| NC score | 0.041525 (rank : 81) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NDB6, Q5JPL2, Q5JPL3, Q5JPL4, Q5JPL5, Q5JPL6, Q68D17, Q6NVY3, Q9UI49 | Gene names | TMEM29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 29. | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.041076 (rank : 82) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.040156 (rank : 83) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
WDR46_MOUSE
|
||||||
| NC score | 0.040100 (rank : 84) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z0H1 | Gene names | Wdr46, Bing4 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.039792 (rank : 85) | θ value | 6.88961 (rank : 162) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.039408 (rank : 86) | θ value | 8.99809 (rank : 182) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
CEP41_HUMAN
|
||||||
| NC score | 0.038981 (rank : 87) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYV8, Q7Z496, Q86TM1, Q8NFU8, Q9H6A3, Q9NPV3 | Gene names | TSGA14, CEP41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.038033 (rank : 88) | θ value | 6.88961 (rank : 164) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.037764 (rank : 89) | θ value | 0.0193708 (rank : 26) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.037131 (rank : 90) | θ value | 0.0193708 (rank : 25) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.036859 (rank : 91) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
EVI2A_MOUSE
|
||||||
| NC score | 0.036363 (rank : 92) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20934 | Gene names | Evi2a, Evi-2, Evi-2a, Evi2 | |||
|
Domain Architecture |

|
|||||
| Description | EVI2A protein precursor (Ecotropic viral integration site 2A protein). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.035166 (rank : 93) | θ value | 6.88961 (rank : 165) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
CP007_MOUSE
|
||||||
| NC score | 0.034864 (rank : 94) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
ILDR1_HUMAN
|
||||||
| NC score | 0.034707 (rank : 95) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86SU0, Q6ZP61, Q7Z578 | Gene names | ILDR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin-like domain-containing receptor 1 precursor. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.034366 (rank : 96) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.034251 (rank : 97) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.033697 (rank : 98) | θ value | 5.27518 (rank : 150) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.033585 (rank : 99) | θ value | 1.38821 (rank : 85) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.032931 (rank : 100) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.032901 (rank : 101) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.032451 (rank : 102) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
RP1_HUMAN
|
||||||
| NC score | 0.032443 (rank : 103) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56715 | Gene names | RP1, ORP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein) (Retinitis pigmentosa 1 protein). | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.031163 (rank : 104) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.029490 (rank : 105) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
FOXC2_HUMAN
|
||||||
| NC score | 0.029302 (rank : 106) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.029208 (rank : 107) | θ value | 5.27518 (rank : 145) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.029082 (rank : 108) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.028724 (rank : 109) | θ value | 1.81305 (rank : 92) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.028419 (rank : 110) | θ value | 5.27518 (rank : 148) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
FOXJ3_HUMAN
|
||||||
| NC score | 0.028169 (rank : 111) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.028068 (rank : 112) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.027901 (rank : 113) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
SPIT1_HUMAN
|
||||||
| NC score | 0.027639 (rank : 114) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.027620 (rank : 115) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
CT075_HUMAN
|
||||||
| NC score | 0.027284 (rank : 116) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WUT4, Q5JWV6, Q9H419 | Gene names | C20orf75 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf75 precursor. | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.026740 (rank : 117) | θ value | 4.03905 (rank : 130) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
TTLL5_MOUSE
|
||||||
| NC score | 0.026682 (rank : 118) | θ value | 1.06291 (rank : 75) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.026564 (rank : 119) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.026296 (rank : 120) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.026275 (rank : 121) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.026166 (rank : 122) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
K1849_MOUSE
|
||||||
| NC score | 0.026125 (rank : 123) | θ value | 1.38821 (rank : 83) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.026058 (rank : 124) | θ value | 6.88961 (rank : 168) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.025666 (rank : 125) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.025274 (rank : 126) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
FOXJ3_MOUSE
|
||||||
| NC score | 0.024962 (rank : 127) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BUR3, Q3UTD8 | Gene names | Foxj3, Kiaa1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.024450 (rank : 128) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.024370 (rank : 129) | θ value | 1.06291 (rank : 73) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
GEMI5_MOUSE
|
||||||
| NC score | 0.023936 (rank : 130) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BX17 | Gene names | Gemin5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
ELOA2_HUMAN
|
||||||
| NC score | 0.023852 (rank : 131) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
FOXC1_MOUSE
|
||||||
| NC score | 0.023668 (rank : 132) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
MEF2D_HUMAN
|
||||||
| NC score | 0.023537 (rank : 133) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14814, Q14815 | Gene names | MEF2D | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
UBP32_HUMAN
|
||||||
| NC score | 0.023465 (rank : 134) | θ value | 0.365318 (rank : 50) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.023003 (rank : 135) | θ value | 1.81305 (rank : 91) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.022003 (rank : 136) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.021962 (rank : 137) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
FOXM1_HUMAN
|
||||||
| NC score | 0.021554 (rank : 138) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.021101 (rank : 139) | θ value | 0.62314 (rank : 58) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
NIPA_MOUSE
|
||||||
| NC score | 0.020489 (rank : 140) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80YV2, Q3TJE6, Q80Z11, Q8BTW5, Q8CI56, Q8R3U7 | Gene names | Zc3hc1, Nipa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear-interacting partner of ALK (Nuclear-interacting partner of anaplastic lymphoma kinase) (mNIPA) (Zinc finger C3HC-type protein 1). | |||||
|
PZRN3_MOUSE
|
||||||
| NC score | 0.020382 (rank : 141) | θ value | 0.813845 (rank : 66) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
DDX24_MOUSE
|
||||||
| NC score | 0.020309 (rank : 142) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
CTTB2_HUMAN
|
||||||
| NC score | 0.020047 (rank : 143) | θ value | 0.62314 (rank : 57) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
GNRP_MOUSE
|
||||||
| NC score | 0.019161 (rank : 144) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27671 | Gene names | Rasgrf1, Cdc25, Grf1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (CDC25Mm). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.018259 (rank : 145) | θ value | 5.27518 (rank : 151) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
LDB3_HUMAN
|
||||||
| NC score | 0.018089 (rank : 146) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
PRGC2_MOUSE
|
||||||
| NC score | 0.018047 (rank : 147) | θ value | 8.99809 (rank : 183) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
RMI1_MOUSE
|
||||||
| NC score | 0.017852 (rank : 148) | θ value | 6.88961 (rank : 166) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D4G9, Q8C560, Q8CI20 | Gene names | Rmi1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RMI1 homolog. | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.017431 (rank : 149) | θ value | 6.88961 (rank : 167) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
ELF2_MOUSE
|
||||||
| NC score | 0.016972 (rank : 150) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
DCX_MOUSE
|
||||||
| NC score | 0.016561 (rank : 151) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
MEF2D_MOUSE
|
||||||
| NC score | 0.016048 (rank : 152) | θ value | 8.99809 (rank : 180) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q63943, Q63944 | Gene names | Mef2d | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.015619 (rank : 153) | θ value | 8.99809 (rank : 184) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
GASP2_HUMAN
|
||||||
| NC score | 0.015600 (rank : 154) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.015349 (rank : 155) | θ value | 5.27518 (rank : 152) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
FRAT1_MOUSE
|
||||||
| NC score | 0.015051 (rank : 156) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70339 | Gene names | Frat1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
ANXA7_MOUSE
|
||||||
| NC score | 0.014907 (rank : 157) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07076 | Gene names | Anxa7, Anx7 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A7 (Annexin VII) (Synexin). | |||||
|
CT075_MOUSE
|
||||||
| NC score | 0.014568 (rank : 158) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59383 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf75 homolog precursor. | |||||
|
PITM1_HUMAN
|
||||||
| NC score | 0.014518 (rank : 159) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.013838 (rank : 160) | θ value | 5.27518 (rank : 144) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
LAP2A_MOUSE
|
||||||
| NC score | 0.013677 (rank : 161) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.013486 (rank : 162) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
GRAM3_MOUSE
|
||||||
| NC score | 0.013255 (rank : 163) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PEM6 | Gene names | Gramd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRAM domain-containing protein 3. | |||||
|
DLGP1_HUMAN
|
||||||
| NC score | 0.012551 (rank : 164) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14490, O14489, P78335 | Gene names | DLGAP1, DAP1, GKAP | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 1 (DAP-1) (Guanylate kinase-associated protein) (hGKAP) (SAP90/PSD-95-associated protein 1) (SAPAP1) (PSD- 95/SAP90-binding protein 1). | |||||
|
MPDZ_MOUSE
|
||||||
| NC score | 0.012474 (rank : 165) | θ value | 8.99809 (rank : 181) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.012474 (rank : 166) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
STAM1_HUMAN
|
||||||
| NC score | 0.012456 (rank : 167) | θ value | 4.03905 (rank : 131) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
UTY_HUMAN
|
||||||
| NC score | 0.012302 (rank : 168) | θ value | 6.88961 (rank : 170) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.011745 (rank : 169) | θ value | 5.27518 (rank : 149) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.010804 (rank : 170) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.010587 (rank : 171) | θ value | 2.36792 (rank : 102) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.010141 (rank : 172) | θ value | 8.99809 (rank : 188) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.009527 (rank : 173) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
UBP2_HUMAN
|
||||||
| NC score | 0.009501 (rank : 174) | θ value | 6.88961 (rank : 169) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.008910 (rank : 175) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.008831 (rank : 176) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.008825 (rank : 177) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.008268 (rank : 178) | θ value | 6.88961 (rank : 163) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
ULK1_HUMAN
|
||||||
| NC score | 0.007449 (rank : 179) | θ value | 0.163984 (rank : 42) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
ROBO3_HUMAN
|
||||||
| NC score | 0.006799 (rank : 180) | θ value | 5.27518 (rank : 147) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
GALT4_MOUSE
|
||||||
| NC score | 0.005793 (rank : 181) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08832, Q3U681 | Gene names | Galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
ZN297_MOUSE
|
||||||
| NC score | 0.005757 (rank : 182) | θ value | 1.06291 (rank : 76) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z0G7 | Gene names | Znf297, Bing1, Zfp297 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 297 (BING1 protein). | |||||
|
TLE3_MOUSE
|
||||||
| NC score | 0.005295 (rank : 183) | θ value | 8.99809 (rank : 187) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q08122, Q923A4 | Gene names | Tle3, Esg | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG) (Grg-3). | |||||
|
NEO1_MOUSE
|
||||||
| NC score | 0.005220 (rank : 184) | θ value | 5.27518 (rank : 146) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97798 | Gene names | Neo1, Ngn | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.005166 (rank : 185) | θ value | 4.03905 (rank : 132) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
NEO1_HUMAN
|
||||||
| NC score | 0.004507 (rank : 186) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92859, O00340 | Gene names | NEO1, NGN | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
PCD12_HUMAN
|
||||||
| NC score | 0.004073 (rank : 187) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NPG4, Q6UXB6, Q96KB8, Q9H7Y6, Q9H8E0 | Gene names | PCDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
MAST3_HUMAN
|
||||||
| NC score | 0.002771 (rank : 188) | θ value | 8.99809 (rank : 179) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
FBN1_HUMAN
|
||||||
| NC score | 0.002037 (rank : 189) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.001841 (rank : 190) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
ALX4_MOUSE
|
||||||
| NC score | 0.001352 (rank : 191) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35137 | Gene names | Alx4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein aristaless-like 4 (ALX-4). | |||||
|
HIPK3_MOUSE
|
||||||
| NC score | 0.000626 (rank : 192) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ERH7, O88906, Q9QZR2 | Gene names | Hipk3, Fist3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeodomain-interacting protein kinase 3 (EC 2.7.11.1) (Fas- interacting serine/threonine-protein kinase) (FIST) (Nuclear body- associated kinase 3) (Nbak3) (Androgen receptor-interacting nuclear protein kinase) (ANPK). | |||||
|
ZN509_MOUSE
|
||||||
| NC score | 0.000499 (rank : 193) | θ value | 1.81305 (rank : 96) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ZFP28_HUMAN
|
||||||
| NC score | -0.001710 (rank : 194) | θ value | 4.03905 (rank : 133) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NHY6, Q9BY30, Q9P2B6 | Gene names | ZFP28, KIAA1431 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 28 homolog (Zfp-28) (Krueppel-like zinc finger factor X6). | |||||