Please be patient as the page loads
|
DOK7_MOUSE
|
||||||
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DOK7_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.931198 (rank : 2) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q18PE1, Q6P6A6, Q86XG5, Q8N2J3, Q8NBC1 | Gene names | DOK7, C4orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
DOK7_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 169 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
DOK6_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 3) | NC score | 0.294941 (rank : 3) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PKX4, Q4V9S3, Q8WUZ8, Q96HI2, Q96LU2 | Gene names | DOK6, DOK5L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 6 (Downstream of tyrosine kinase 6). | |||||
|
DOK4_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 4) | NC score | 0.270350 (rank : 6) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TEW6, O75209, Q9BTP2, Q9NVV3 | Gene names | DOK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
DOK4_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 5) | NC score | 0.269859 (rank : 7) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99KE3, Q3U0X3 | Gene names | Dok4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
DOK5_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 6) | NC score | 0.271383 (rank : 4) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9P104, Q5T7Y0, Q5TE53, Q8TEW7, Q96H13, Q9BZ24, Q9NQF4, Q9Y411 | Gene names | DOK5, C20orf180 | |||
|
Domain Architecture |
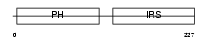
|
|||||
| Description | Docking protein 5 (Downstream of tyrosine kinase 5) (IRS6) (Protein dok-5). | |||||
|
DOK5_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 7) | NC score | 0.270934 (rank : 5) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91ZM9, Q8BRI3, Q9CSM6 | Gene names | Dok5 | |||
|
Domain Architecture |
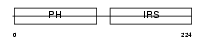
|
|||||
| Description | Docking protein 5 (Downstream of tyrosine kinase 5) (Protein dok-5). | |||||
|
DEND_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 8) | NC score | 0.111622 (rank : 18) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
ZN598_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 9) | NC score | 0.092361 (rank : 22) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
TBX21_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 10) | NC score | 0.044720 (rank : 79) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 11) | NC score | 0.090380 (rank : 23) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
FRS3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 12) | NC score | 0.225209 (rank : 9) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91WJ0 | Gene names | Frs3, Frs2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (FRS2-beta). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 13) | NC score | 0.045049 (rank : 78) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
FRS3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.227030 (rank : 8) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43559, Q5T3D5 | Gene names | FRS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (Suc1-associated neurotrophic factor target 2) (SNT-2) (FGFR signaling adaptor SNT2). | |||||
|
SF3B4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.052230 (rank : 57) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
DOK3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.210136 (rank : 10) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QZK7 | Gene names | Dok3, Dokl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 3 (Downstream of tyrosine kinase 3) (P62(dok)-like protein) (DOK-L). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 17) | NC score | 0.096030 (rank : 21) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.097434 (rank : 20) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.043042 (rank : 86) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.089370 (rank : 25) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
AATK_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.016430 (rank : 150) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80YE4, O35211, Q3U2U5, Q3UHR8, Q66JN3, Q80YE3, Q8CB63, Q8CHE2 | Gene names | Aatk, Kiaa0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase). | |||||
|
FRS2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.199382 (rank : 11) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C180 | Gene names | Frs2, Frs2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT) (FRS2 alpha). | |||||
|
SOX10_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.051606 (rank : 59) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.086727 (rank : 26) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
FRS2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.197260 (rank : 12) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WU20, O43558, Q7LDQ6 | Gene names | FRS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT). | |||||
|
SOX10_HUMAN
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.051007 (rank : 61) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.031333 (rank : 108) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
DVL1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.048332 (rank : 68) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14640 | Gene names | DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.042037 (rank : 89) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.059358 (rank : 42) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.025456 (rank : 122) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
ABI2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.034582 (rank : 102) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
MAST3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.010867 (rank : 163) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
RFIP5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.047693 (rank : 74) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R361 | Gene names | Rab11fip5, D6Ertd32e, Rip11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11). | |||||
|
RREB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.016568 (rank : 149) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
TOIP2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.047896 (rank : 73) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
ZBT45_HUMAN
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.009837 (rank : 167) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96K62 | Gene names | ZBTB45, ZNF499 | |||
|
Domain Architecture |
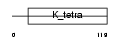
|
|||||
| Description | Zinc finger and BTB domain-containing protein 45 (Zinc finger protein 499). | |||||
|
DOK3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.181359 (rank : 14) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7L591, Q8N864, Q9BQB3, Q9H666 | Gene names | DOK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 3 (Downstream of tyrosine kinase 3). | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.043261 (rank : 84) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.024276 (rank : 125) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.098111 (rank : 19) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.037100 (rank : 96) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.090211 (rank : 24) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CAMLG_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.067836 (rank : 32) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49069 | Gene names | CAMLG, CAML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium signal-modulating cyclophilin ligand (CAML). | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.056989 (rank : 48) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.077769 (rank : 29) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
DOK2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.168546 (rank : 17) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60496 | Gene names | DOK2 | |||
|
Domain Architecture |
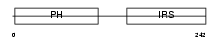
|
|||||
| Description | Docking protein 2 (Downstream of tyrosine kinase 2) (p56(dok-2)). | |||||
|
HSPB8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.065747 (rank : 33) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJY1, Q6FIH3, Q9UKS3 | Gene names | HSPB8, CRYAC, E2IG1, HSP22 | |||
|
Domain Architecture |
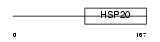
|
|||||
| Description | Heat-shock protein beta-8 (HspB8) (Alpha crystallin C chain) (Small stress protein-like protein HSP22) (E2-induced gene 1 protein) (Protein kinase H11). | |||||
|
KLF13_MOUSE
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.007622 (rank : 173) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JJZ6, Q9ESX3, Q9JHF8 | Gene names | Klf13, Bteb3, Fklf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 13 (Transcription factor BTEB3) (Basic transcription element-binding protein 3) (BTE-binding protein 3) (RANTES factor of late activated T-lymphocytes 1) (RFLAT-1) (Erythroid transcription factor FKLF-2). | |||||
|
LRC56_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.044060 (rank : 81) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IYG6, Q8N3Q4 | Gene names | LRRC56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 56. | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.062385 (rank : 38) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.069028 (rank : 31) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
DOK1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.185987 (rank : 13) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99704, O43204, Q9UHG6 | Gene names | DOK1 | |||
|
Domain Architecture |
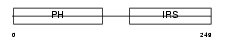
|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)) (pp62). | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.045210 (rank : 76) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.041398 (rank : 91) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
HXA5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.011100 (rank : 162) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09021, Q91VQ5 | Gene names | Hoxa5, Hox-1.3, Hoxa-5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A5 (Hox-1.3) (M2). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.048060 (rank : 71) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TRIP6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.021470 (rank : 135) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.059906 (rank : 40) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.047997 (rank : 72) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
CDT1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.042536 (rank : 88) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H211, Q86XX9, Q96CJ5, Q96GK5, Q96H67, Q96HE6, Q9BWM0 | Gene names | CDT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP). | |||||
|
EGF_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.010309 (rank : 166) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01132, Q6P9J2 | Gene names | Egf | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor]. | |||||
|
HNF3B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.022774 (rank : 130) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35583, Q60602 | Gene names | Foxa2, Hnf3b, Tcf-3b, Tcf3b | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-beta (HNF-3B) (Forkhead box protein A2). | |||||
|
M3K5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.005450 (rank : 180) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99683, Q5THN3, Q99461 | Gene names | MAP3K5, ASK1, MAPKKK5, MEKK5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 5 (EC 2.7.11.25) (MAPK/ERK kinase kinase 5) (MEK kinase 5) (MEKK 5) (Apoptosis signal- regulating kinase 1) (ASK-1). | |||||
|
PRRT1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.065578 (rank : 34) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35449 | Gene names | Prrt1, Ng5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.051751 (rank : 58) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SYGP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.020807 (rank : 136) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.033269 (rank : 103) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.036302 (rank : 97) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.047044 (rank : 75) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
PDCD7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.038419 (rank : 95) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N8D1, Q96AK8, Q9Y6D7 | Gene names | PDCD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18) (HES18). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.021896 (rank : 132) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.029443 (rank : 112) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.032372 (rank : 105) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.045208 (rank : 77) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
UTX_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.026994 (rank : 119) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15550 | Gene names | UTX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed X chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the X chromosome). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.058485 (rank : 45) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.021550 (rank : 134) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.028343 (rank : 113) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
DOK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.173881 (rank : 16) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P97465, Q9R213 | Gene names | Dok1, Dok | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)). | |||||
|
DVL2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.043787 (rank : 82) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
FOXE3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.017988 (rank : 145) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QY14 | Gene names | Foxe3 | |||
|
Domain Architecture |
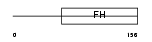
|
|||||
| Description | Forkhead box protein E3. | |||||
|
GLIS3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.007646 (rank : 172) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.026383 (rank : 121) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
PAX4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.010788 (rank : 164) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P32115, P97341, Q9QUR1, Q9R091 | Gene names | Pax4, Pax-4 | |||
|
Domain Architecture |
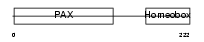
|
|||||
| Description | Paired box protein Pax-4. | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.053560 (rank : 54) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.035118 (rank : 100) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.035682 (rank : 99) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SOX18_HUMAN
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.034831 (rank : 101) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35713, Q9NPH8 | Gene names | SOX18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.054657 (rank : 52) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.015763 (rank : 152) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.011547 (rank : 161) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CCD96_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.018071 (rank : 144) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CN004_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.031459 (rank : 107) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
DOK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.174239 (rank : 15) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O70469, O70272, Q99KL1 | Gene names | Dok2, Frip | |||
|
Domain Architecture |
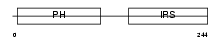
|
|||||
| Description | Docking protein 2 (Downstream of tyrosine kinase 2) (p56(dok-2)) (Dok- related protein) (Dok-R) (IL-four receptor-interacting protein) (FRIP). | |||||
|
GAK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.009129 (rank : 169) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
GRIN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.044534 (rank : 80) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60269, Q5SVF0 | Gene names | GPRIN2, KIAA0514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 2 (GRIN2). | |||||
|
IRX1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.017217 (rank : 148) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78414, Q7Z2F8, Q8N312 | Gene names | IRX1, IRXA1 | |||
|
Domain Architecture |
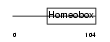
|
|||||
| Description | Iroquois-class homeodomain protein IRX-1 (Iroquois homeobox protein 1) (Homeodomain protein IRXA1). | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.030593 (rank : 110) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
NPDC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.032973 (rank : 104) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQX5, Q9BTD6, Q9BXT3, Q9NQS2, Q9Y434 | Gene names | NPDC1 | |||
|
Domain Architecture |
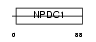
|
|||||
| Description | Neural proliferation differentiation and control protein 1 precursor (NPDC-1 protein). | |||||
|
PRRT1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.056079 (rank : 49) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99946, Q96DW3, Q96NQ8 | Gene names | PRRT1, C6orf31, NG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
RUSC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.026998 (rank : 118) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BVN2, Q9UPY4, Q9Y4T5 | Gene names | RUSC1, NESCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1 (New molecule containing SH3 at the carboxy-terminus) (Nesca). | |||||
|
SF01_MOUSE
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.039534 (rank : 93) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
SNPC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.042852 (rank : 87) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91XA5, Q3UK17, Q8CBS9 | Gene names | Snapc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2). | |||||
|
TR240_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.024490 (rank : 123) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHV7, O60334 | Gene names | THRAP1, ARC250, KIAA0593, TRAP240 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor-associated protein complex 240 kDa component (Trap240) (Thyroid hormone receptor-associated protein 1) (Vitamin D3 receptor-interacting protein complex component DRIP250) (DRIP 250) (Activator-recruited cofactor 250 kDa component) (ARC250). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.026970 (rank : 120) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
ZN414_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.027345 (rank : 117) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96IQ9 | Gene names | ZNF414 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 414. | |||||
|
BORG3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.031075 (rank : 109) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NZY7, Q8TB51 | Gene names | CDC42EP5, BORG3, CEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 5 (Binder of Rho GTPases 3). | |||||
|
COBA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.027953 (rank : 116) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
DAZP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.020166 (rank : 138) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JII5 | Gene names | Dazap1 | |||
|
Domain Architecture |
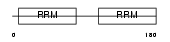
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.014790 (rank : 156) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
FZD8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.008898 (rank : 170) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H461 | Gene names | FZD8 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-8 precursor (Fz-8) (hFz8). | |||||
|
HNF3A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.021654 (rank : 133) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35582, Q61108 | Gene names | Foxa1, Hnf3a, Tcf-3a, Tcf3a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-alpha (HNF-3A) (Forkhead box protein A1). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.064758 (rank : 35) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MCL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.024215 (rank : 126) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97287, Q3TUS0, Q792P0, Q9CRI4 | Gene names | Mcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog (Bcl-2-related protein EAT/mcl1). | |||||
|
NMDE4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.010605 (rank : 165) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.058968 (rank : 44) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RP3A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.015760 (rank : 153) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.024469 (rank : 124) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.048143 (rank : 69) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.051310 (rank : 60) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.050540 (rank : 64) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
BNIPL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.015464 (rank : 154) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z465, Q6DK43, Q8TCY7, Q8WYG2 | Gene names | BNIPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2/adenovirus E1B 19 kDa-interacting protein 2-like protein. | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.028304 (rank : 114) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
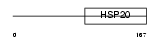
HSPB8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.052643 (rank : 55) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JK92 | Gene names | Hspb8, Cryac, Hsp22 | |||
|
Domain Architecture |
|
|||||
| Description | Heat-shock protein beta-8 (HspB8) (Alpha crystallin C chain) (Small stress protein-like protein HSP22). | |||||
|
INSM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.006308 (rank : 176) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 683 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01101 | Gene names | INSM1, IA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.024169 (rank : 127) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
LZTS2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.016219 (rank : 151) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
MARCS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.039850 (rank : 92) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.043099 (rank : 85) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.020768 (rank : 137) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.057174 (rank : 47) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.043441 (rank : 83) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.012572 (rank : 160) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.041669 (rank : 90) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.019387 (rank : 142) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
DAZP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.017785 (rank : 147) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |
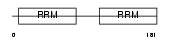
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
DEND_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.078234 (rank : 28) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80TS7 | Gene names | Ddn, Kiaa0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
HGS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.022096 (rank : 131) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HNF3A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.019765 (rank : 140) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55317, Q9H2A0 | Gene names | FOXA1, HNF3A, TCF3A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-alpha (HNF-3A) (Forkhead box protein A1). | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.013108 (rank : 157) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
LATS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.004687 (rank : 181) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.031651 (rank : 106) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
NANO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.019775 (rank : 139) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80WY3, Q3UTS9, Q8BIJ9 | Gene names | Nanos1, Nos | |||
|
Domain Architecture |

|
|||||
| Description | Nanos homolog 1. | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.038745 (rank : 94) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
RERE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.036166 (rank : 98) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SORC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.012611 (rank : 159) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96PQ0, Q9P2L7 | Gene names | SORCS2, KIAA1329 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
T22D4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.023133 (rank : 129) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.048124 (rank : 70) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CABP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.005560 (rank : 179) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CNTRB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.008451 (rank : 171) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
DUOX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.006964 (rank : 174) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRD9, Q6ZMB3, Q6ZR09, Q9NZC1 | Gene names | DUOX1, DUOX, LNOX1, THOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 1 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH thyroid oxidase 1) (Thyroid oxidase 1) (Large NOX 1) (Long NOX 1). | |||||
|
ERF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.009203 (rank : 168) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
GDF11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.006187 (rank : 177) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1W4, Q9QX55, Q9R221 | Gene names | Gdf11, Bmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Growth/differentiation factor 11 precursor (GDF-11) (Bone morphogenetic protein 11). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.029608 (rank : 111) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HNF3B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.017981 (rank : 146) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y261, Q8WUW4, Q96DF7 | Gene names | FOXA2, HNF3B, TCF3B | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-beta (HNF-3B) (Forkhead box protein A2). | |||||
|
HXD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.006640 (rank : 175) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P10628, P97451, Q3UXL6 | Gene names | Hoxd4, Hox-4.2, Hoxd-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D4 (Hox-4.2) (Hox-5.1). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.019666 (rank : 141) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.028028 (rank : 115) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
K1849_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.018344 (rank : 143) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
KLF4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.003010 (rank : 182) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 694 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60793, P70421 | Gene names | Klf4, Ezf, Gklf, Zie | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 4 (Gut-enriched krueppel-like factor) (Epithelial zinc-finger protein EZF). | |||||
|
LAMB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.005866 (rank : 178) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
MK04_MOUSE
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.002040 (rank : 183) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P5G0 | Gene names | Mapk4, Erk4, Prkm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 4 (EC 2.7.11.24) (Extracellular signal-regulated kinase 4) (ERK-4). | |||||
|
NHS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.023721 (rank : 128) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6T4R5, Q5J7Q0, Q5J7Q1, Q68DR5 | Gene names | NHS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nance-Horan syndrome protein (Congenital cataracts and dental anomalies protein). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.012622 (rank : 158) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.048461 (rank : 67) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
SMG5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.015073 (rank : 155) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
ST32C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.001696 (rank : 184) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86UX6, Q5T0Q5, Q86UE1 | Gene names | STK32C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 32C (EC 2.7.11.1) (PKE) (YANK3). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.063575 (rank : 37) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.050779 (rank : 62) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.063844 (rank : 36) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.052239 (rank : 56) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CEL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.050089 (rank : 65) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.050606 (rank : 63) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.054825 (rank : 51) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
MISS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.059339 (rank : 43) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.082556 (rank : 27) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.060845 (rank : 39) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.059384 (rank : 41) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.055278 (rank : 50) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.070517 (rank : 30) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SC24C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.057587 (rank : 46) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SON_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.050001 (rank : 66) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.053696 (rank : 53) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
DOK7_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 169 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
DOK7_HUMAN
|
||||||
| NC score | 0.931198 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q18PE1, Q6P6A6, Q86XG5, Q8N2J3, Q8NBC1 | Gene names | DOK7, C4orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
DOK6_HUMAN
|
||||||
| NC score | 0.294941 (rank : 3) | θ value | 9.29e-05 (rank : 3) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PKX4, Q4V9S3, Q8WUZ8, Q96HI2, Q96LU2 | Gene names | DOK6, DOK5L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 6 (Downstream of tyrosine kinase 6). | |||||
|
DOK5_HUMAN
|
||||||
| NC score | 0.271383 (rank : 4) | θ value | 0.00390308 (rank : 6) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9P104, Q5T7Y0, Q5TE53, Q8TEW7, Q96H13, Q9BZ24, Q9NQF4, Q9Y411 | Gene names | DOK5, C20orf180 | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 5 (Downstream of tyrosine kinase 5) (IRS6) (Protein dok-5). | |||||
|
DOK5_MOUSE
|
||||||
| NC score | 0.270934 (rank : 5) | θ value | 0.00390308 (rank : 7) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91ZM9, Q8BRI3, Q9CSM6 | Gene names | Dok5 | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 5 (Downstream of tyrosine kinase 5) (Protein dok-5). | |||||
|
DOK4_HUMAN
|
||||||
| NC score | 0.270350 (rank : 6) | θ value | 0.00390308 (rank : 4) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TEW6, O75209, Q9BTP2, Q9NVV3 | Gene names | DOK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
DOK4_MOUSE
|
||||||
| NC score | 0.269859 (rank : 7) | θ value | 0.00390308 (rank : 5) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99KE3, Q3U0X3 | Gene names | Dok4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
FRS3_HUMAN
|
||||||
| NC score | 0.227030 (rank : 8) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43559, Q5T3D5 | Gene names | FRS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (Suc1-associated neurotrophic factor target 2) (SNT-2) (FGFR signaling adaptor SNT2). | |||||
|
FRS3_MOUSE
|
||||||
| NC score | 0.225209 (rank : 9) | θ value | 0.0148317 (rank : 12) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91WJ0 | Gene names | Frs3, Frs2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (FRS2-beta). | |||||
|
DOK3_MOUSE
|
||||||
| NC score | 0.210136 (rank : 10) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QZK7 | Gene names | Dok3, Dokl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 3 (Downstream of tyrosine kinase 3) (P62(dok)-like protein) (DOK-L). | |||||
|
FRS2_MOUSE
|
||||||
| NC score | 0.199382 (rank : 11) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C180 | Gene names | Frs2, Frs2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT) (FRS2 alpha). | |||||
|
FRS2_HUMAN
|
||||||
| NC score | 0.197260 (rank : 12) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WU20, O43558, Q7LDQ6 | Gene names | FRS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT). | |||||
|
DOK1_HUMAN
|
||||||
| NC score | 0.185987 (rank : 13) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99704, O43204, Q9UHG6 | Gene names | DOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)) (pp62). | |||||
|
DOK3_HUMAN
|
||||||
| NC score | 0.181359 (rank : 14) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7L591, Q8N864, Q9BQB3, Q9H666 | Gene names | DOK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 3 (Downstream of tyrosine kinase 3). | |||||
|
DOK2_MOUSE
|
||||||
| NC score | 0.174239 (rank : 15) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O70469, O70272, Q99KL1 | Gene names | Dok2, Frip | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 2 (Downstream of tyrosine kinase 2) (p56(dok-2)) (Dok- related protein) (Dok-R) (IL-four receptor-interacting protein) (FRIP). | |||||
|
DOK1_MOUSE
|
||||||
| NC score | 0.173881 (rank : 16) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P97465, Q9R213 | Gene names | Dok1, Dok | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)). | |||||
|
DOK2_HUMAN
|
||||||
| NC score | 0.168546 (rank : 17) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60496 | Gene names | DOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 2 (Downstream of tyrosine kinase 2) (p56(dok-2)). | |||||
|
DEND_HUMAN
|
||||||
| NC score | 0.111622 (rank : 18) | θ value | 0.00509761 (rank : 8) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.098111 (rank : 19) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.097434 (rank : 20) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.096030 (rank : 21) | θ value | 0.0330416 (rank : 17) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
ZN598_HUMAN
|
||||||
| NC score | 0.092361 (rank : 22) | θ value | 0.00509761 (rank : 9) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.090380 (rank : 23) | θ value | 0.00869519 (rank : 11) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.090211 (rank : 24) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.089370 (rank : 25) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.086727 (rank : 26) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.082556 (rank : 27) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
DEND_MOUSE
|
||||||
| NC score | 0.078234 (rank : 28) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80TS7 | Gene names | Ddn, Kiaa0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.077769 (rank : 29) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.070517 (rank : 30) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.069028 (rank : 31) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
CAMLG_HUMAN
|
||||||
| NC score | 0.067836 (rank : 32) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49069 | Gene names | CAMLG, CAML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium signal-modulating cyclophilin ligand (CAML). | |||||
|
HSPB8_HUMAN
|
||||||
| NC score | 0.065747 (rank : 33) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJY1, Q6FIH3, Q9UKS3 | Gene names | HSPB8, CRYAC, E2IG1, HSP22 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-8 (HspB8) (Alpha crystallin C chain) (Small stress protein-like protein HSP22) (E2-induced gene 1 protein) (Protein kinase H11). | |||||
|
PRRT1_MOUSE
|
||||||
| NC score | 0.065578 (rank : 34) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35449 | Gene names | Prrt1, Ng5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.064758 (rank : 35) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.063844 (rank : 36) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.063575 (rank : 37) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.062385 (rank : 38) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.060845 (rank : 39) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.059906 (rank : 40) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.059384 (rank : 41) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.059358 (rank : 42) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.059339 (rank : 43) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.058968 (rank : 44) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.058485 (rank : 45) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.057587 (rank : 46) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.057174 (rank : 47) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.056989 (rank : 48) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
PRRT1_HUMAN
|
||||||
| NC score | 0.056079 (rank : 49) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99946, Q96DW3, Q96NQ8 | Gene names | PRRT1, C6orf31, NG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.055278 (rank : 50) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.054825 (rank : 51) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.054657 (rank : 52) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.053696 (rank : 53) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.053560 (rank : 54) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
HSPB8_MOUSE
|
||||||
| NC score | 0.052643 (rank : 55) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JK92 | Gene names | Hspb8, Cryac, Hsp22 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-8 (HspB8) (Alpha crystallin C chain) (Small stress protein-like protein HSP22). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.052239 (rank : 56) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
SF3B4_HUMAN
|
||||||
| NC score | 0.052230 (rank : 57) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.051751 (rank : 58) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SOX10_MOUSE
|
||||||
| NC score | 0.051606 (rank : 59) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.051310 (rank : 60) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
SOX10_HUMAN
|
||||||
| NC score | 0.051007 (rank : 61) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.050779 (rank : 62) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.050606 (rank : 63) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.050540 (rank : 64) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.050089 (rank : 65) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.050001 (rank : 66) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.048461 (rank : 67) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
DVL1_HUMAN
|
||||||
| NC score | 0.048332 (rank : 68) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14640 | Gene names | DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.048143 (rank : 69) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.048124 (rank : 70) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.048060 (rank : 71) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.047997 (rank : 72) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
TOIP2_MOUSE
|
||||||
| NC score | 0.047896 (rank : 73) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
RFIP5_MOUSE
|
||||||
| NC score | 0.047693 (rank : 74) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R361 | Gene names | Rab11fip5, D6Ertd32e, Rip11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11). | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.047044 (rank : 75) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.045210 (rank : 76) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.045208 (rank : 77) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.045049 (rank : 78) | θ value | 0.0193708 (rank : 13) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
TBX21_HUMAN
|
||||||
| NC score | 0.044720 (rank : 79) | θ value | 0.00665767 (rank : 10) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
GRIN2_HUMAN
|
||||||
| NC score | 0.044534 (rank : 80) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60269, Q5SVF0 | Gene names | GPRIN2, KIAA0514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 2 (GRIN2). | |||||
|
LRC56_HUMAN
|
||||||
| NC score | 0.044060 (rank : 81) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IYG6, Q8N3Q4 | Gene names | LRRC56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 56. | |||||
|
DVL2_MOUSE
|
||||||
| NC score | 0.043787 (rank : 82) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.043441 (rank : 83) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.043261 (rank : 84) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.043099 (rank : 85) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.043042 (rank : 86) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SNPC2_MOUSE
|
||||||
| NC score | 0.042852 (rank : 87) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91XA5, Q3UK17, Q8CBS9 | Gene names | Snapc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2). | |||||
|
CDT1_HUMAN
|
||||||
| NC score | 0.042536 (rank : 88) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H211, Q86XX9, Q96CJ5, Q96GK5, Q96H67, Q96HE6, Q9BWM0 | Gene names | CDT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.042037 (rank : 89) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.041669 (rank : 90) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.041398 (rank : 91) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.039850 (rank : 92) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
SF01_MOUSE
|
||||||
| NC score | 0.039534 (rank : 93) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.038745 (rank : 94) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
PDCD7_HUMAN
|
||||||
| NC score | 0.038419 (rank : 95) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N8D1, Q96AK8, Q9Y6D7 | Gene names | PDCD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18) (HES18). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.037100 (rank : 96) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.036302 (rank : 97) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.036166 (rank : 98) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.035682 (rank : 99) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.035118 (rank : 100) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
SOX18_HUMAN
|
||||||
| NC score | 0.034831 (rank : 101) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35713, Q9NPH8 | Gene names | SOX18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
ABI2_HUMAN
|
||||||
| NC score | 0.034582 (rank : 102) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.033269 (rank : 103) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
NPDC1_HUMAN
|
||||||
| NC score | 0.032973 (rank : 104) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQX5, Q9BTD6, Q9BXT3, Q9NQS2, Q9Y434 | Gene names | NPDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Neural proliferation differentiation and control protein 1 precursor (NPDC-1 protein). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.032372 (rank : 105) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.031651 (rank : 106) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
CN004_HUMAN
|
||||||
| NC score | 0.031459 (rank : 107) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.031333 (rank : 108) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
BORG3_HUMAN
|
||||||
| NC score | 0.031075 (rank : 109) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NZY7, Q8TB51 | Gene names | CDC42EP5, BORG3, CEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 5 (Binder of Rho GTPases 3). | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.030593 (rank : 110) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.029608 (rank : 111) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.029443 (rank : 112) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.028343 (rank : 113) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.028304 (rank : 114) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.028028 (rank : 115) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
COBA1_HUMAN
|
||||||
| NC score | 0.027953 (rank : 116) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
ZN414_HUMAN
|
||||||
| NC score | 0.027345 (rank : 117) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96IQ9 | Gene names | ZNF414 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 414. | |||||
|
RUSC1_HUMAN
|
||||||
| NC score | 0.026998 (rank : 118) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BVN2, Q9UPY4, Q9Y4T5 | Gene names | RUSC1, NESCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1 (New molecule containing SH3 at the carboxy-terminus) (Nesca). | |||||
|
UTX_HUMAN
|
||||||
| NC score | 0.026994 (rank : 119) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15550 | Gene names | UTX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed X chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the X chromosome). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.026970 (rank : 120) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.026383 (rank : 121) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.025456 (rank : 122) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
TR240_HUMAN
|
||||||
| NC score | 0.024490 (rank : 123) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHV7, O60334 | Gene names | THRAP1, ARC250, KIAA0593, TRAP240 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor-associated protein complex 240 kDa component (Trap240) (Thyroid hormone receptor-associated protein 1) (Vitamin D3 receptor-interacting protein complex component DRIP250) (DRIP 250) (Activator-recruited cofactor 250 kDa component) (ARC250). | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.024469 (rank : 124) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.024276 (rank : 125) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MCL1_MOUSE
|
||||||
| NC score | 0.024215 (rank : 126) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97287, Q3TUS0, Q792P0, Q9CRI4 | Gene names | Mcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog (Bcl-2-related protein EAT/mcl1). | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.024169 (rank : 127) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
NHS_HUMAN
|
||||||
| NC score | 0.023721 (rank : 128) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6T4R5, Q5J7Q0, Q5J7Q1, Q68DR5 | Gene names | NHS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nance-Horan syndrome protein (Congenital cataracts and dental anomalies protein). | |||||
|
T22D4_MOUSE
|
||||||
| NC score | 0.023133 (rank : 129) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
HNF3B_MOUSE
|
||||||
| NC score | 0.022774 (rank : 130) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35583, Q60602 | Gene names | Foxa2, Hnf3b, Tcf-3b, Tcf3b | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-beta (HNF-3B) (Forkhead box protein A2). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.022096 (rank : 131) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.021896 (rank : 132) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
HNF3A_MOUSE
|
||||||
| NC score | 0.021654 (rank : 133) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35582, Q61108 | Gene names | Foxa1, Hnf3a, Tcf-3a, Tcf3a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-alpha (HNF-3A) (Forkhead box protein A1). | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.021550 (rank : 134) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
TRIP6_HUMAN
|
||||||
| NC score | 0.021470 (rank : 135) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
SYGP1_HUMAN
|
||||||
| NC score | 0.020807 (rank : 136) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.020768 (rank : 137) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
DAZP1_MOUSE
|
||||||
| NC score | 0.020166 (rank : 138) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JII5 | Gene names | Dazap1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
NANO1_MOUSE
|
||||||
| NC score | 0.019775 (rank : 139) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80WY3, Q3UTS9, Q8BIJ9 | Gene names | Nanos1, Nos | |||
|
Domain Architecture |

|
|||||
| Description | Nanos homolog 1. | |||||
|
HNF3A_HUMAN
|
||||||
| NC score | 0.019765 (rank : 140) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55317, Q9H2A0 | Gene names | FOXA1, HNF3A, TCF3A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-alpha (HNF-3A) (Forkhead box protein A1). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.019666 (rank : 141) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.019387 (rank : 142) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
K1849_HUMAN
|
||||||
| NC score | 0.018344 (rank : 143) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
CCD96_HUMAN
|
||||||
| NC score | 0.018071 (rank : 144) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
FOXE3_MOUSE
|
||||||
| NC score | 0.017988 (rank : 145) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QY14 | Gene names | Foxe3 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E3. | |||||
|
HNF3B_HUMAN
|
||||||
| NC score | 0.017981 (rank : 146) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y261, Q8WUW4, Q96DF7 | Gene names | FOXA2, HNF3B, TCF3B | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-beta (HNF-3B) (Forkhead box protein A2). | |||||
|
DAZP1_HUMAN
|
||||||
| NC score | 0.017785 (rank : 147) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
IRX1_HUMAN
|
||||||
| NC score | 0.017217 (rank : 148) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78414, Q7Z2F8, Q8N312 | Gene names | IRX1, IRXA1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-1 (Iroquois homeobox protein 1) (Homeodomain protein IRXA1). | |||||
|
RREB1_HUMAN
|
||||||
| NC score | 0.016568 (rank : 149) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
AATK_MOUSE
|
||||||
| NC score | 0.016430 (rank : 150) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80YE4, O35211, Q3U2U5, Q3UHR8, Q66JN3, Q80YE3, Q8CB63, Q8CHE2 | Gene names | Aatk, Kiaa0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase). | |||||
|
LZTS2_MOUSE
|
||||||
| NC score | 0.016219 (rank : 151) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.015763 (rank : 152) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
RP3A_HUMAN
|
||||||
| NC score | 0.015760 (rank : 153) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
BNIPL_HUMAN
|
||||||
| NC score | 0.015464 (rank : 154) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z465, Q6DK43, Q8TCY7, Q8WYG2 | Gene names | BNIPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2/adenovirus E1B 19 kDa-interacting protein 2-like protein. | |||||
|
SMG5_HUMAN
|
||||||
| NC score | 0.015073 (rank : 155) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.014790 (rank : 156) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.013108 (rank : 157) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.012622 (rank : 158) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
SORC2_HUMAN
|
||||||
| NC score | 0.012611 (rank : 159) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96PQ0, Q9P2L7 | Gene names | SORCS2, KIAA1329 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.012572 (rank : 160) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.011547 (rank : 161) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
HXA5_MOUSE
|
||||||
| NC score | 0.011100 (rank : 162) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09021, Q91VQ5 | Gene names | Hoxa5, Hox-1.3, Hoxa-5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A5 (Hox-1.3) (M2). | |||||
|
MAST3_HUMAN
|
||||||
| NC score | 0.010867 (rank : 163) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
PAX4_MOUSE
|
||||||
| NC score | 0.010788 (rank : 164) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P32115, P97341, Q9QUR1, Q9R091 | Gene names | Pax4, Pax-4 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-4. | |||||
|
NMDE4_HUMAN
|
||||||
| NC score | 0.010605 (rank : 165) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
EGF_MOUSE
|
||||||
| NC score | 0.010309 (rank : 166) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01132, Q6P9J2 | Gene names | Egf | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor]. | |||||
|
ZBT45_HUMAN
|
||||||
| NC score | 0.009837 (rank : 167) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96K62 | Gene names | ZBTB45, ZNF499 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 45 (Zinc finger protein 499). | |||||
|
ERF_MOUSE
|
||||||
| NC score | 0.009203 (rank : 168) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
GAK_HUMAN
|
||||||
| NC score | 0.009129 (rank : 169) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
FZD8_HUMAN
|
||||||
| NC score | 0.008898 (rank : 170) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H461 | Gene names | FZD8 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-8 precursor (Fz-8) (hFz8). | |||||
|
CNTRB_HUMAN
|
||||||
| NC score | 0.008451 (rank : 171) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
GLIS3_MOUSE
|
||||||
| NC score | 0.007646 (rank : 172) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
KLF13_MOUSE
|
||||||
| NC score | 0.007622 (rank : 173) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JJZ6, Q9ESX3, Q9JHF8 | Gene names | Klf13, Bteb3, Fklf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 13 (Transcription factor BTEB3) (Basic transcription element-binding protein 3) (BTE-binding protein 3) (RANTES factor of late activated T-lymphocytes 1) (RFLAT-1) (Erythroid transcription factor FKLF-2). | |||||
|
DUOX1_HUMAN
|
||||||
| NC score | 0.006964 (rank : 174) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRD9, Q6ZMB3, Q6ZR09, Q9NZC1 | Gene names | DUOX1, DUOX, LNOX1, THOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 1 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH thyroid oxidase 1) (Thyroid oxidase 1) (Large NOX 1) (Long NOX 1). | |||||
|
HXD4_MOUSE
|
||||||
| NC score | 0.006640 (rank : 175) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P10628, P97451, Q3UXL6 | Gene names | Hoxd4, Hox-4.2, Hoxd-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D4 (Hox-4.2) (Hox-5.1). | |||||
|
INSM1_HUMAN
|
||||||
| NC score | 0.006308 (rank : 176) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 683 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01101 | Gene names | INSM1, IA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
GDF11_MOUSE
|
||||||
| NC score | 0.006187 (rank : 177) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1W4, Q9QX55, Q9R221 | Gene names | Gdf11, Bmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Growth/differentiation factor 11 precursor (GDF-11) (Bone morphogenetic protein 11). | |||||
|
LAMB2_MOUSE
|
||||||
| NC score | 0.005866 (rank : 178) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
CABP4_MOUSE
|
||||||
| NC score | 0.005560 (rank : 179) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
M3K5_HUMAN
|
||||||
| NC score | 0.005450 (rank : 180) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99683, Q5THN3, Q99461 | Gene names | MAP3K5, ASK1, MAPKKK5, MEKK5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 5 (EC 2.7.11.25) (MAPK/ERK kinase kinase 5) (MEK kinase 5) (MEKK 5) (Apoptosis signal- regulating kinase 1) (ASK-1). | |||||
|
LATS1_HUMAN
|
||||||
| NC score | 0.004687 (rank : 181) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
KLF4_MOUSE
|
||||||
| NC score | 0.003010 (rank : 182) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 694 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60793, P70421 | Gene names | Klf4, Ezf, Gklf, Zie | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 4 (Gut-enriched krueppel-like factor) (Epithelial zinc-finger protein EZF). | |||||
|
MK04_MOUSE
|
||||||
| NC score | 0.002040 (rank : 183) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P5G0 | Gene names | Mapk4, Erk4, Prkm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 4 (EC 2.7.11.24) (Extracellular signal-regulated kinase 4) (ERK-4). | |||||
|
ST32C_HUMAN
|
||||||
| NC score | 0.001696 (rank : 184) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86UX6, Q5T0Q5, Q86UE1 | Gene names | STK32C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 32C (EC 2.7.11.1) (PKE) (YANK3). | |||||