Please be patient as the page loads
|
SORC2_HUMAN
|
||||||
| SwissProt Accessions | Q96PQ0, Q9P2L7 | Gene names | SORCS2, KIAA1329 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SORC1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.980588 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WY21, Q59GG7, Q5JVT7, Q5JVT8, Q5VY14, Q86WQ1, Q86WQ2, Q9H1Y1, Q9H1Y2 | Gene names | SORCS1, SORCS | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (hSorCS). | |||||
|
SORC1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.979600 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JLC4, Q8VI45, Q922I1, Q9QY21 | Gene names | Sorcs1, Sorcs | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (mSorCS). | |||||
|
SORC2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96PQ0, Q9P2L7 | Gene names | SORCS2, KIAA1329 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SORC2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.992881 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9EPR5 | Gene names | Sorcs2 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SORC3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.976781 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
SORC3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.978401 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VI51, Q9CTR4 | Gene names | Sorcs3 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
SORL_MOUSE
|
||||||
| θ value | 2.17053e-70 (rank : 7) | NC score | 0.465750 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
SORL_HUMAN
|
||||||
| θ value | 3.13423e-69 (rank : 8) | NC score | 0.462021 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92673, Q92856 | Gene names | SORL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (SorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11). | |||||
|
SORT_HUMAN
|
||||||
| θ value | 6.32992e-62 (rank : 9) | NC score | 0.871421 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99523, Q8IZ49 | Gene names | SORT1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin precursor (Neurotensin receptor 3) (NTR3) (NT3) (Glycoprotein 95) (Gp95) (100 kDa NT receptor). | |||||
|
SORT_MOUSE
|
||||||
| θ value | 1.19377e-60 (rank : 10) | NC score | 0.871880 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PHU5, Q3UHE2, Q8K043, Q9QXW6 | Gene names | Sort1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sortilin precursor (Neurotensin receptor 3) (NTR3) (mNTR3). | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 11) | NC score | 0.027533 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 12) | NC score | 0.023048 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 13) | NC score | 0.024833 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 14) | NC score | 0.030539 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.043416 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
SPS2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.030936 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99611, Q9BUQ2 | Gene names | SEPHS2, SPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Selenide, water dikinase 2 (EC 2.7.9.3) (Selenophosphate synthetase 2) (Selenium donor protein 2). | |||||
|
WDR46_MOUSE
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.035761 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z0H1 | Gene names | Wdr46, Bing4 | |||
|
Domain Architecture |
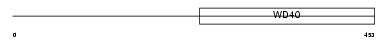
|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.025573 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.052288 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
FOXH1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.021610 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
PUNC_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.020367 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BQC3, O70246, Q792T2, Q9Z2S6 | Gene names | Punc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
TBX15_MOUSE
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.012848 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70306, O54840 | Gene names | Tbx15, Tbx14, Tbx8 | |||
|
Domain Architecture |
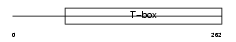
|
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15) (MmTBx8). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.026473 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
AAPK1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.003118 (rank : 81) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13131, O00286, Q5D0E1, Q9UNQ4 | Gene names | PRKAA1, AMPK1 | |||
|
Domain Architecture |
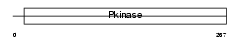
|
|||||
| Description | 5'-AMP-activated protein kinase catalytic subunit alpha-1 (EC 2.7.11.1) (AMPK alpha-1 chain). | |||||
|
AAPK1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.003131 (rank : 80) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5EG47 | Gene names | Prkaa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-AMP-activated protein kinase catalytic subunit alpha-1 (EC 2.7.11.1) (AMPK alpha-1 chain). | |||||
|
LTBP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.025187 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
NEK3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.002125 (rank : 82) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R0A5, Q9Z0X9 | Gene names | Nek3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek3 (EC 2.7.11.1) (NimA-related protein kinase 3). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.011631 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.004811 (rank : 78) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.014573 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
TNR16_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.021538 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z0W1 | Gene names | Ngfr, Tnfrsf16 | |||
|
Domain Architecture |
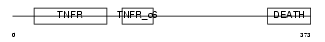
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Low affinity neurotrophin receptor p75NTR). | |||||
|
IRX5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.010873 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JKQ4, Q80WV4, Q9JLL5 | Gene names | Irx5, Irxb2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.011796 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
TBX21_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.011607 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
ZAR1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.024255 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
ITB5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.008357 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70309, O70308, O88347 | Gene names | Itgb5 | |||
|
Domain Architecture |
|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
NOS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.009279 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29475 | Gene names | NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
SMOO_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.014226 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.011413 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
GLI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | -0.000308 (rank : 85) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.010260 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
DRP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.008321 (rank : 71) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.004814 (rank : 77) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.009585 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.011198 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PLOD3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.010744 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0E1, Q542E0, Q9CYY9 | Gene names | Plod3 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 precursor (EC 1.14.11.4) (Lysyl hydroxylase 3) (LH3). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.011908 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
PSG7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.005136 (rank : 76) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13046, Q15232 | Gene names | PSG7 | |||
|
Domain Architecture |
|
|||||
| Description | Pregnancy-specific beta-1-glycoprotein 7 precursor (PSBG-7). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.008745 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.011374 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SEN54_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.015217 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z6J9, Q86WV3, Q86XE4, Q8N9H2 | Gene names | TSEN54, SEN54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen54 (tRNA-intron endonuclease Sen54) (HsSen54). | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.012122 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
CUL7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.024519 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.005374 (rank : 75) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.004539 (rank : 79) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
TNR16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.019329 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08138 | Gene names | NGFR, TNFRSF16 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Gp80-LNGFR) (p75 ICD) (Low affinity neurotrophin receptor p75NTR) (CD271 antigen). | |||||
|
WDR46_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.024625 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15213, Q5STK5 | Gene names | WDR46, BING4, C6orf11 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
DMRTD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.010956 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
DOK7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.012611 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.008883 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
TLK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.001118 (rank : 83) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86UE8, Q9UKI7, Q9Y4F7 | Gene names | TLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2) (PKU-alpha). | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.013572 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
FBX41_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.006987 (rank : 73) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.007647 (rank : 72) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.005727 (rank : 74) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.000613 (rank : 84) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
CD320_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.068771 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPF0, Q53HF7 | Gene names | CD320, 8D6A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD320 antigen precursor (8D6 antigen) (FDC-signaling molecule 8D6) (FDC-SM-8D6). | |||||
|
LDLR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.051795 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01130, Q53ZD9 | Gene names | LDLR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor precursor (LDL receptor). | |||||
|
LDLR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.053326 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35951 | Gene names | Ldlr | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor precursor (LDL receptor). | |||||
|
LRP12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.052606 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
LRP1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.050932 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.051665 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.053681 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
LRP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.058027 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
LRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.053551 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |
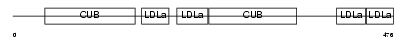
|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
LRP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.050369 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75096 | Gene names | LRP4, KIAA0816, MEGF7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 4 precursor (Multiple epidermal growth factor-like domains 7). | |||||
|
LRP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.050399 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VI56, Q8BPX5, Q8CBB3, Q8CCP5 | Gene names | Lrp4, Kiaa0816 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 4 precursor (LDLR dan). | |||||
|
LRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.052208 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75197, Q96TD6, Q9UP66 | Gene names | LRP5, LRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor. | |||||
|
LRP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.051946 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91VN0, O88883, Q9R208 | Gene names | Lrp5, Lrp7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor (LRP7) (Lr3). | |||||
|
LRP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.052879 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75581 | Gene names | LRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LRP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.053225 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88572 | Gene names | Lrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LRP8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.053167 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14114, O14968, Q86V27, Q99876, Q9BR78 | Gene names | LRP8, APOER2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
LRP8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.053552 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q924X6, Q8CAK9, Q8CDF5, Q921B6 | Gene names | Lrp8, Apoer2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
VLDLR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.054354 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98155, Q5VVF6 | Gene names | VLDLR | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
VLDLR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.056497 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98156, Q64022 | Gene names | Vldlr | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
SORC2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96PQ0, Q9P2L7 | Gene names | SORCS2, KIAA1329 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SORC2_MOUSE
|
||||||
| NC score | 0.992881 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9EPR5 | Gene names | Sorcs2 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SORC1_HUMAN
|
||||||
| NC score | 0.980588 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WY21, Q59GG7, Q5JVT7, Q5JVT8, Q5VY14, Q86WQ1, Q86WQ2, Q9H1Y1, Q9H1Y2 | Gene names | SORCS1, SORCS | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (hSorCS). | |||||
|
SORC1_MOUSE
|
||||||
| NC score | 0.979600 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JLC4, Q8VI45, Q922I1, Q9QY21 | Gene names | Sorcs1, Sorcs | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (mSorCS). | |||||
|
SORC3_MOUSE
|
||||||
| NC score | 0.978401 (rank : 5) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VI51, Q9CTR4 | Gene names | Sorcs3 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
SORC3_HUMAN
|
||||||
| NC score | 0.976781 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
SORT_MOUSE
|
||||||
| NC score | 0.871880 (rank : 7) | θ value | 1.19377e-60 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PHU5, Q3UHE2, Q8K043, Q9QXW6 | Gene names | Sort1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sortilin precursor (Neurotensin receptor 3) (NTR3) (mNTR3). | |||||
|
SORT_HUMAN
|
||||||
| NC score | 0.871421 (rank : 8) | θ value | 6.32992e-62 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99523, Q8IZ49 | Gene names | SORT1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin precursor (Neurotensin receptor 3) (NTR3) (NT3) (Glycoprotein 95) (Gp95) (100 kDa NT receptor). | |||||
|
SORL_MOUSE
|
||||||
| NC score | 0.465750 (rank : 9) | θ value | 2.17053e-70 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
SORL_HUMAN
|
||||||
| NC score | 0.462021 (rank : 10) | θ value | 3.13423e-69 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92673, Q92856 | Gene names | SORL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (SorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11). | |||||
|
CD320_HUMAN
|
||||||
| NC score | 0.068771 (rank : 11) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPF0, Q53HF7 | Gene names | CD320, 8D6A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD320 antigen precursor (8D6 antigen) (FDC-signaling molecule 8D6) (FDC-SM-8D6). | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.058027 (rank : 12) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
VLDLR_MOUSE
|
||||||
| NC score | 0.056497 (rank : 13) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98156, Q64022 | Gene names | Vldlr | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
VLDLR_HUMAN
|
||||||
| NC score | 0.054354 (rank : 14) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98155, Q5VVF6 | Gene names | VLDLR | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
LRP1_HUMAN
|
||||||
| NC score | 0.053681 (rank : 15) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
LRP8_MOUSE
|
||||||
| NC score | 0.053552 (rank : 16) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q924X6, Q8CAK9, Q8CDF5, Q921B6 | Gene names | Lrp8, Apoer2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
LRP3_HUMAN
|
||||||
| NC score | 0.053551 (rank : 17) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
LDLR_MOUSE
|
||||||
| NC score | 0.053326 (rank : 18) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35951 | Gene names | Ldlr | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor precursor (LDL receptor). | |||||
|
LRP6_MOUSE
|
||||||
| NC score | 0.053225 (rank : 19) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88572 | Gene names | Lrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LRP8_HUMAN
|
||||||
| NC score | 0.053167 (rank : 20) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14114, O14968, Q86V27, Q99876, Q9BR78 | Gene names | LRP8, APOER2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
LRP6_HUMAN
|
||||||
| NC score | 0.052879 (rank : 21) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75581 | Gene names | LRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LRP12_MOUSE
|
||||||
| NC score | 0.052606 (rank : 22) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.052288 (rank : 23) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
LRP5_HUMAN
|
||||||
| NC score | 0.052208 (rank : 24) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75197, Q96TD6, Q9UP66 | Gene names | LRP5, LRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor. | |||||
|
LRP5_MOUSE
|
||||||
| NC score | 0.051946 (rank : 25) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91VN0, O88883, Q9R208 | Gene names | Lrp5, Lrp7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor (LRP7) (Lr3). | |||||
|
LDLR_HUMAN
|
||||||
| NC score | 0.051795 (rank : 26) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01130, Q53ZD9 | Gene names | LDLR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor precursor (LDL receptor). | |||||
|
LRP1B_MOUSE
|
||||||
| NC score | 0.051665 (rank : 27) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP1B_HUMAN
|
||||||
| NC score | 0.050932 (rank : 28) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP4_MOUSE
|
||||||
| NC score | 0.050399 (rank : 29) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VI56, Q8BPX5, Q8CBB3, Q8CCP5 | Gene names | Lrp4, Kiaa0816 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 4 precursor (LDLR dan). | |||||
|
LRP4_HUMAN
|
||||||
| NC score | 0.050369 (rank : 30) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75096 | Gene names | LRP4, KIAA0816, MEGF7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 4 precursor (Multiple epidermal growth factor-like domains 7). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.043416 (rank : 31) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
WDR46_MOUSE
|
||||||
| NC score | 0.035761 (rank : 32) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z0H1 | Gene names | Wdr46, Bing4 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
SPS2_HUMAN
|
||||||
| NC score | 0.030936 (rank : 33) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99611, Q9BUQ2 | Gene names | SEPHS2, SPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Selenide, water dikinase 2 (EC 2.7.9.3) (Selenophosphate synthetase 2) (Selenium donor protein 2). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.030539 (rank : 34) | θ value | 0.0431538 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.027533 (rank : 35) | θ value | 0.0113563 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.026473 (rank : 36) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.025573 (rank : 37) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
LTBP2_HUMAN
|
||||||
| NC score | 0.025187 (rank : 38) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.024833 (rank : 39) | θ value | 0.0431538 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
WDR46_HUMAN
|
||||||
| NC score | 0.024625 (rank : 40) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15213, Q5STK5 | Gene names | WDR46, BING4, C6orf11 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
CUL7_HUMAN
|
||||||
| NC score | 0.024519 (rank : 41) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
ZAR1_HUMAN
|
||||||
| NC score | 0.024255 (rank : 42) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.023048 (rank : 43) | θ value | 0.0193708 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
FOXH1_HUMAN
|
||||||
| NC score | 0.021610 (rank : 44) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
TNR16_MOUSE
|
||||||
| NC score | 0.021538 (rank : 45) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z0W1 | Gene names | Ngfr, Tnfrsf16 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Low affinity neurotrophin receptor p75NTR). | |||||
|
PUNC_MOUSE
|
||||||
| NC score | 0.020367 (rank : 46) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BQC3, O70246, Q792T2, Q9Z2S6 | Gene names | Punc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
TNR16_HUMAN
|
||||||
| NC score | 0.019329 (rank : 47) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08138 | Gene names | NGFR, TNFRSF16 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Gp80-LNGFR) (p75 ICD) (Low affinity neurotrophin receptor p75NTR) (CD271 antigen). | |||||
|
SEN54_HUMAN
|
||||||
| NC score | 0.015217 (rank : 48) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z6J9, Q86WV3, Q86XE4, Q8N9H2 | Gene names | TSEN54, SEN54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen54 (tRNA-intron endonuclease Sen54) (HsSen54). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.014573 (rank : 49) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.014226 (rank : 50) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.013572 (rank : 51) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
TBX15_MOUSE
|
||||||
| NC score | 0.012848 (rank : 52) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70306, O54840 | Gene names | Tbx15, Tbx14, Tbx8 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15) (MmTBx8). | |||||
|
DOK7_MOUSE
|
||||||
| NC score | 0.012611 (rank : 53) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.012122 (rank : 54) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.011908 (rank : 55) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.011796 (rank : 56) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.011631 (rank : 57) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TBX21_HUMAN
|
||||||
| NC score | 0.011607 (rank : 58) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.011413 (rank : 59) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.011374 (rank : 60) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.011198 (rank : 61) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
DMRTD_HUMAN
|
||||||
| NC score | 0.010956 (rank : 62) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
IRX5_MOUSE
|
||||||
| NC score | 0.010873 (rank : 63) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JKQ4, Q80WV4, Q9JLL5 | Gene names | Irx5, Irxb2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2). | |||||
|
PLOD3_MOUSE
|
||||||
| NC score | 0.010744 (rank : 64) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0E1, Q542E0, Q9CYY9 | Gene names | Plod3 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 precursor (EC 1.14.11.4) (Lysyl hydroxylase 3) (LH3). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.010260 (rank : 65) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.009585 (rank : 66) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NOS1_HUMAN
|
||||||
| NC score | 0.009279 (rank : 67) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29475 | Gene names | NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.008883 (rank : 68) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.008745 (rank : 69) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
ITB5_MOUSE
|
||||||
| NC score | 0.008357 (rank : 70) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70309, O70308, O88347 | Gene names | Itgb5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
DRP2_HUMAN
|
||||||
| NC score | 0.008321 (rank : 71) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.007647 (rank : 72) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
FBX41_MOUSE
|
||||||
| NC score | 0.006987 (rank : 73) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.005727 (rank : 74) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.005374 (rank : 75) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PSG7_HUMAN
|
||||||
| NC score | 0.005136 (rank : 76) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13046, Q15232 | Gene names | PSG7 | |||
|
Domain Architecture |

|
|||||
| Description | Pregnancy-specific beta-1-glycoprotein 7 precursor (PSBG-7). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.004814 (rank : 77) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.004811 (rank : 78) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.004539 (rank : 79) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
AAPK1_MOUSE
|
||||||
| NC score | 0.003131 (rank : 80) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5EG47 | Gene names | Prkaa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-AMP-activated protein kinase catalytic subunit alpha-1 (EC 2.7.11.1) (AMPK alpha-1 chain). | |||||
|
AAPK1_HUMAN
|
||||||
| NC score | 0.003118 (rank : 81) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13131, O00286, Q5D0E1, Q9UNQ4 | Gene names | PRKAA1, AMPK1 | |||
|
Domain Architecture |

|
|||||
| Description | 5'-AMP-activated protein kinase catalytic subunit alpha-1 (EC 2.7.11.1) (AMPK alpha-1 chain). | |||||
|
NEK3_MOUSE
|
||||||
| NC score | 0.002125 (rank : 82) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R0A5, Q9Z0X9 | Gene names | Nek3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek3 (EC 2.7.11.1) (NimA-related protein kinase 3). | |||||
|
TLK2_HUMAN
|
||||||
| NC score | 0.001118 (rank : 83) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86UE8, Q9UKI7, Q9Y4F7 | Gene names | TLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2) (PKU-alpha). | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.000613 (rank : 84) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
GLI1_HUMAN
|
||||||
| NC score | -0.000308 (rank : 85) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||